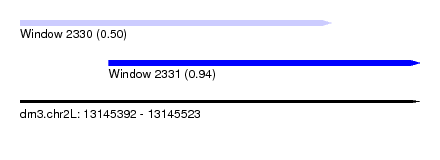
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,145,392 – 13,145,523 |
| Length | 131 |
| Max. P | 0.942204 |

| Location | 13,145,392 – 13,145,494 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.58375 |
| G+C content | 0.49623 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -12.57 |
| Energy contribution | -12.74 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 13145392 102 + 23011544 -----------UUUCGUAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CGCCCGACC--UGGUCAAAAGCGUACCGUCC- -----------....(((((...((((((((...(((.....((((((....))))))(((((((....))))))))))((...--....))..)--)))))))..)).))).....- ( -25.00, z-score = 0.81, R) >droAna3.scaffold_12916 3886150 106 - 16180835 -----------UUUUGGAGCCACUUGAUUAGCACGCUCUGAGCCAAGUUUAAACGUGGGCUGCUUAACCAAACGGCAGCCGACGACCAUCCAUCCAACCCUCCAAGUCAAAGCUAGC- -----------.(((((((..(((((.((((......))))..))))).....(((.(((((((.........))))))).)))...............)))))))...........- ( -30.40, z-score = -1.99, R) >droEre2.scaffold_4929 14346120 102 - 26641161 -----------UUUCGUAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CACCCGACC--UGGUCAAAAGCGUACCGUCC- -----------....(((((...((((((((...(((.....((((((....))))))(((((((....))))))))))((...--....))..)--)))))))..)).))).....- ( -25.00, z-score = 0.40, R) >droYak2.chr2L 9568352 103 + 22324452 -----------UUUCGUAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CACCCGACC--UGGUCAAAAGCGAACCGUCCA -----------.(((((......((((((((...(((.....((((((....))))))(((((((....))))))))))((...--....))..)--)))))))..)))))....... ( -24.40, z-score = 0.58, R) >droSec1.super_16 1300236 102 + 1878335 -----------UUUCGUAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGUCGAUA--CACCCGACC--UGGUCAAAAGCGUAGCGUCC- -----------...(((.(((..((((((((...........((((((....))))))(((((((....))))))).((((...--....)))))--)))))))..).)).)))...- ( -27.60, z-score = -0.19, R) >droSim1.chr2L 12939038 102 + 22036055 -----------UUUCGUAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGUCGAUA--CACCCGACC--UGGUCAAAAGCGUAGCGUCC- -----------...(((.(((..((((((((...........((((((....))))))(((((((....))))))).((((...--....)))))--)))))))..).)).)))...- ( -27.60, z-score = -0.19, R) >droPer1.super_46 383436 92 - 590583 -----------UUUUGGACCUACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCAGCCACUA--CACCAGGCC--CGACCAGACC----------- -----------.(((((.....((((..(((...(((.....((((((....))))))(((((((....))))))))))..)))--...))))..--...)))))..----------- ( -22.60, z-score = -0.65, R) >dp4.chr4_group4 854588 92 + 6586962 -----------UUUUGGACCUACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCAGCCACUA--CACCAGGCC--CGACCAGACC----------- -----------.(((((.....((((..(((...(((.....((((((....))))))(((((((....))))))))))..)))--...))))..--...)))))..----------- ( -22.60, z-score = -0.65, R) >droWil1.scaffold_180708 6328145 110 - 12563649 -UUUUUUUUUCUUUUGGGCCUAGUUGAUUAGCACGCUUUGAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGACUGCCCGU---CAGGCAGUCAGAGCCAAAUAGAGAGAAC---- -...((((((((....(((((.(((((.((((.((((((((((...))))))).))).)))).))))).....(((((((...---..))))))))).)))....)))))))).---- ( -39.80, z-score = -4.14, R) >droVir3.scaffold_12963 13190502 92 - 20206255 ----UUUUUUUUUGUCGAGCUACUUGAUUAGCACGCUCUUAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCGGGCAAUACAAAAACGGCAC---------------------- ----........(((((.((((......))))..((((....((((((....))))))(((((((....)))))))))))..........))))).---------------------- ( -29.00, z-score = -3.06, R) >droMoj3.scaffold_6500 6142792 96 - 32352404 UGUCUUUUUUUUUUUUGAGCUACUUGAUUAGCACGCUCUUAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCGGGCAAUACAAAAACGGCAC---------------------- ((((.(((((........((((......))))..((((....((((((....))))))(((((((....))))))))))).....))))).)))).---------------------- ( -27.20, z-score = -2.36, R) >droGri2.scaffold_15252 7015882 82 - 17193109 --------------UCGAGGUACUUGAUUAGCACGCUCUUAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGUGAGCCAAACAAAAACGGCAC---------------------- --------------..(((((.((.....)).)).)))....((((((....))))))(((((((....)))))))..(((..........)))..---------------------- ( -19.40, z-score = -0.38, R) >consensus ___________UUUUGGAGCCACUUGAUUAGCACGCUCUGAGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCAGCCGAUA__CACCCGGCC__UGGUCAAAAGCG_A_C_____ ..........................................((((((....))))))(((((((....))))))).......................................... (-12.57 = -12.74 + 0.17)

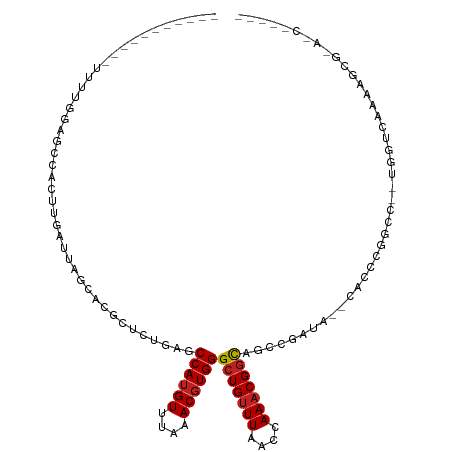
| Location | 13,145,421 – 13,145,523 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 60.08 |
| Shannon entropy | 0.79215 |
| G+C content | 0.49154 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13145421 102 + 23011544 AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CGCCCGACCUGG--UCAAAAGCGUACCGUCCAA-UAGCAGCGAUGGCAACAAAAAGAGCUC----- ..((((((....))))))((((((..(((....)))...((.((--(((..(((...)--))....))))).)).....-.))))))..((....))..........----- ( -28.10, z-score = -0.25, R) >droAna3.scaffold_12916 3886179 112 - 16180835 AGCCAAGUUUAAACGUGGGCUGCUUAACCAAACGGCAGCCGACGACCAUCCAUCCAACCCUCCAAGUCAAAGCUAGCCAUAGCUGGCUAUAGAAGCAACAAAAAAAGCGCUC ......((((...(((.(((((((.........))))))).)))..........................(((((((....)))))))....))))................ ( -29.20, z-score = -1.84, R) >droEre2.scaffold_4929 14346149 102 - 26641161 AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CACCCGACCUGG--UCAAAAGCGUACCGUCCAA-UAACAGCGAUGGCAACAACAAGAGCUC----- ..((((((....))))))((((((.......((((..((.....--.(((......))--).....))...))))....-.))))))(.((....)).)........----- ( -24.10, z-score = 0.37, R) >droYak2.chr2L 9568381 103 + 22324452 AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA--CACCCGACCUGG--UCAAAAGCGAACCGUCCAAAUAGCAGCGAUGGCAGCAACAACAGCUC----- .((((((((......(((((.((((.(((....))).((.....--.(((......))--).....)))))).)))))......))).)))))(((.......))).----- ( -27.40, z-score = -0.17, R) >droSec1.super_16 1300265 78 + 1878335 AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGUCGAUA--CACCCGACCUGG--UCAAAAGCGUAGCGUCCAA-UAC----------------------------- .((.((((((.......((((....))))....(((.((((...--....))))...)--))..)))))).))......-...----------------------------- ( -19.00, z-score = -0.08, R) >droSim1.chr2L 12939067 78 + 22036055 AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGUCGAUA--CACCCGACCUGG--UCAAAAGCGUAGCGUCCAA-UAC----------------------------- .((.((((((.......((((....))))....(((.((((...--....))))...)--))..)))))).))......-...----------------------------- ( -19.00, z-score = -0.08, R) >droWil1.scaffold_180708 6328184 99 - 12563649 AGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGACUGCCCGU---CAGGCAGUCAGAGCCAAAUAGAGAGAACAUCAGCUAAAGACCACACUAAAAGACUU---------- ((((((((....)))))((((((((....))))(((((((...---..)))))))..)))).................))).....................---------- ( -25.30, z-score = -2.19, R) >droVir3.scaffold_12963 13190538 89 - 20206255 AGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGCGGGCAAUACAAAAACGGCACAUCAGCUGCAAAAGCAACAACAGCAACAACAAC----------------------- ..((((((....))))))((((((..........((.(............).))......(((.....)))...)))))).........----------------------- ( -19.70, z-score = -0.66, R) >droGri2.scaffold_15252 7015908 80 - 17193109 AGCCAUGUUUAAACGUGGGCUGUUUAACCAAACGGUGAGCCAAACAAAAACGGCACAUAAGCUACAACAAAAACAACAAU-------------------------------- ..((((((....))))))(((((((....)))))))..(((..........)))..........................-------------------------------- ( -15.50, z-score = -0.85, R) >consensus AGCCAUGUUUAAACGUGGGCUGUUUAGCCAAACGGCAGCCGAUA__CACCCGACCUGG__UCAAAAGCGAAACGUCCAA_UAC_AGC_A____A__A_______________ ..((((((....))))))(((((((....)))))))............................................................................ (-12.12 = -12.36 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:03 2011