| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,142,862 – 13,142,932 |
| Length | 70 |
| Max. P | 0.784677 |

| Location | 13,142,862 – 13,142,932 |
|---|---|
| Length | 70 |
| Sequences | 9 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 67.57 |
| Shannon entropy | 0.65981 |
| G+C content | 0.43923 |
| Mean single sequence MFE | -18.27 |
| Consensus MFE | -6.89 |
| Energy contribution | -7.86 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
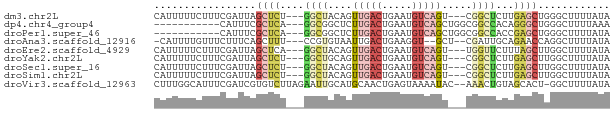
>dm3.chr2L 13142862 70 + 23011544 CAUUUUUCUUUCGAUUAGCUCU---GGCUACAGUUGACUGAAUGUCAGU---CGGCUCUUGAGCUGGGCUUUUAUA ................(((.(.---.(((..(((((((((.....))))---)))))....)))..))))...... ( -23.00, z-score = -3.33, R) >dp4.chr4_group4 850622 62 + 6586962 -----------CAUUUCGCUCA---GGCGGCUCUUGACUGAAUGUCAGCUGGCGGCCACAGGGCUGGGCUUUUAAA -----------......((((.---(.(((((...(((.....)))))))).)((((....))))))))....... ( -21.00, z-score = -0.96, R) >droPer1.super_46 379190 62 - 590583 -----------CAUUUCGCUCA---GGCGGCUCUUGACUGAAUGUCAGCUGGCGGCCACCGAGCUGGGCUUUUAUA -----------......((((.---(((.(((.(((((.....)))))..))).)))...))))............ ( -20.30, z-score = -0.76, R) >droAna3.scaffold_12916 3884283 68 - 16180835 -CAUUUUGUUUCUUUCAGCUUU---CCGUGUAAUUGACUGAAGGU--GCU--CGAUUGCAGAACCAGGCUUUUAUA -...............(((((.---...((((((((((.......--).)--)))))))).....)))))...... ( -11.00, z-score = 0.49, R) >droEre2.scaffold_4929 14343497 70 - 26641161 CAUUUUUCUUUCGAUUAGCUCA---GGCUACAGUUGACUGAAUGUCAGU---UGGUUCUUUAGCUUGGCUUUUAUA ................(((.((---(((((.(((..((((.....))))---..)..)).))))))))))...... ( -19.30, z-score = -3.02, R) >droYak2.chr2L 9565670 70 + 22324452 CAUUUUUCUUUCGAUUAGCUCU---GGCUGCAGUUGACUGAAUGUCAGU---CGGCUCUUGAGCUUGGCUUUUAUA ................((((..---((((..(((((((((.....))))---)))))....)))).))))...... ( -19.60, z-score = -1.73, R) >droSec1.super_16 1297672 70 + 1878335 CAUUUUUCUUUCGAUUAGCUCU---GGCUACAGUUGACUGAAUGUCAGU---CGGCUCUUGAGCUUGGCUUUUAUA ................((((..---((((..(((((((((.....))))---)))))....)))).))))...... ( -20.70, z-score = -2.76, R) >droSim1.chr2L 12936474 70 + 22036055 CAUUUUUCUUUCGAUUAGCUCU---GGCUACAGUUGACUGAAUGUCAGU---CGGCUCUUGAGCUUGGCUUUUAUA ................((((..---((((..(((((((((.....))))---)))))....)))).))))...... ( -20.70, z-score = -2.76, R) >droVir3.scaffold_12963 7011251 73 + 20206255 CUUUGGCAUUUCGAUCGUGUCUUAGAAUUGCAUGCAACUGAGUAAAAUAC--AAACUGUAGCACU-GGCUUUUAUA ((..(((((.......)))))..))((..((.(((.((..(((.......--..))))).)))..-.))..))... ( -8.80, z-score = 1.72, R) >consensus CAUUUUUCUUUCGAUUAGCUCU___GGCUGCAGUUGACUGAAUGUCAGU___CGGCUCUUGAGCUUGGCUUUUAUA .................((((....((((....(((((.....))))).....))))...))))............ ( -6.89 = -7.86 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:37:01 2011