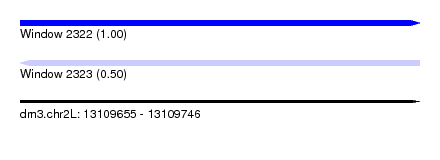
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,109,655 – 13,109,746 |
| Length | 91 |
| Max. P | 0.996779 |

| Location | 13,109,655 – 13,109,746 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.50064 |
| G+C content | 0.33737 |
| Mean single sequence MFE | -18.41 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.06 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
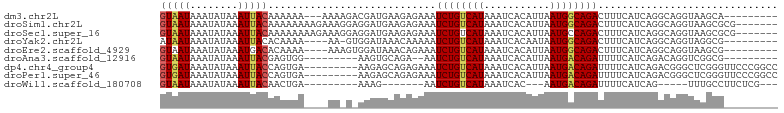
>dm3.chr2L 13109655 91 + 23011544 GUAAUAAAUAUAAAUUACAAAAAA---AAAAGACGAUGAAGAGAAAUCUGUCAUAAAUCACAUUAAUGGCAGACUUUCAUCAGGCAGGUAAGCA--------- (((((........)))))......---.......(((((.(((...((((((((.(((...))).))))))))))))))))..((......)).--------- ( -17.70, z-score = -3.67, R) >droSim1.chr2L 12901872 96 + 22036055 GUAAUAAAUAUAAAUUACAAAAAAAAGAAAGGAGGAUGAAGAGAAAUCUGUCAUAAAUCACAUUAAUGGCAGACUUUCAUCAGGCAGGUAAGCGCG------- (((((........)))))................(((((.(((...((((((((.(((...))).))))))))))))))))..((......))...------- ( -17.40, z-score = -2.78, R) >droSec1.super_16 1264893 96 + 1878335 GUAAUAAAUAUAAAUUACAAAAAAAAGAAAGGAGGAUGAAGAGAAAUCUGUCAUAAAUCACAUUAAUGCCAGACUUUCAUCAGGCAGGUAAGCGCG------- (((((........))))).............((..(((((((....))).))))...))...(((.((((.((......)).))))..))).....------- ( -15.10, z-score = -1.53, R) >droYak2.chr2L 9531594 89 + 22324452 AUAAUAAAUAUAAAUUACACAAAA----AA-GUGGAUAAACAAAAAUCUGUCAUAAAUCACAAUAAUGGCAGACUUUCAUCAGGCAGGUAGGCG--------- .................(((....----..-)))............((((((((...........))))))))..........((......)).--------- ( -11.30, z-score = -1.70, R) >droEre2.scaffold_4929 14307030 90 - 26641161 GUAAUAAAUAUAAAUGACACAAAA----AAAGUGGAUAAACAGAAAUCUGUCAUAAAUCACAUUAAUGGCAGACUUUCAUCAGGCAGGUAAGCG--------- ..............((((((....----...)))........((((((((((((.(((...))).)))))))).)))).))).((......)).--------- ( -15.00, z-score = -2.31, R) >droAna3.scaffold_12916 3843231 83 - 16180835 GUAAUAAAUAUAAAUUACGAGUGG---------AAGUGCAGA--AAUCUGUCAUAAAUCACAUUAAUGACAGAUUUUCAUCAGACAGGUCGGCG--------- (((((........)))))..((.(---------(..(((.((--((((((((((.(((...))).)))))))))))).....).))..)).)).--------- ( -18.80, z-score = -2.83, R) >dp4.chr4_group4 814312 94 + 6586962 GUGAUAAAUAUAAAUUACCAGUGA---------AAGAGCAGAGAAAUCUGUCAUAAAUCACAUUAAUGACAGAUUUUCAUCAGACGGGCUCGGGUUCCCGGCC .((((...............((..---------....)).(((((.((((((((.(((...))).)))))))))))))))))....(((.(((....)))))) ( -23.90, z-score = -2.31, R) >droPer1.super_46 343006 94 - 590583 GUGAUAAAUAUAAAUUACCAGUGA---------AAGAGCAGAGAAAUCUGUCAUAAAUCACAUUAAUGACAGAUUUUCAUCAGACGGGCUCGGGUUCCCGGCC .((((...............((..---------....)).(((((.((((((((.(((...))).)))))))))))))))))....(((.(((....)))))) ( -23.90, z-score = -2.31, R) >droWil1.scaffold_180708 6183671 76 + 12563649 GUAAUAAAUAUAAAUUACAACUGA---------AAAG-------AAUCUGUCAUAAAUCAC---AAUGACAGAUUUUCAUCAG-----UUUGCCUUCUCG--- (((((........)))))((((((---------..((-------((((((((((.......---.))))))))))))..))))-----))..........--- ( -22.60, z-score = -6.86, R) >consensus GUAAUAAAUAUAAAUUACAAAAAA____AA_G_AGAUGAAGAGAAAUCUGUCAUAAAUCACAUUAAUGACAGACUUUCAUCAGGCAGGUAAGCG_________ (((((........)))))............................((((((((...........)))))))).............................. ( -9.07 = -9.06 + -0.01)

| Location | 13,109,655 – 13,109,746 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.50064 |
| G+C content | 0.33737 |
| Mean single sequence MFE | -16.82 |
| Consensus MFE | -6.14 |
| Energy contribution | -6.11 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
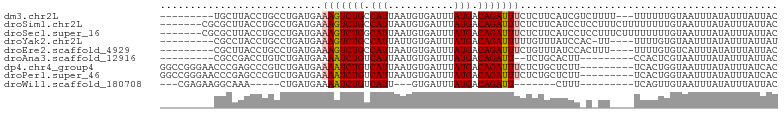
>dm3.chr2L 13109655 91 - 23011544 ---------UGCUUACCUGCCUGAUGAAAGUCUGCCAUUAAUGUGAUUUAUGACAGAUUUCUCUUCAUCGUCUUUU---UUUUUUGUAAUUUAUAUUUAUUAC ---------.((......))..((((((((((((.(((...........))).))))))....)))))).......---......(((((........))))) ( -13.10, z-score = -1.44, R) >droSim1.chr2L 12901872 96 - 22036055 -------CGCGCUUACCUGCCUGAUGAAAGUCUGCCAUUAAUGUGAUUUAUGACAGAUUUCUCUUCAUCCUCCUUUCUUUUUUUUGUAAUUUAUAUUUAUUAC -------...((......))..((((((((((((.(((...........))).))))))....))))))................(((((........))))) ( -12.50, z-score = -1.43, R) >droSec1.super_16 1264893 96 - 1878335 -------CGCGCUUACCUGCCUGAUGAAAGUCUGGCAUUAAUGUGAUUUAUGACAGAUUUCUCUUCAUCCUCCUUUCUUUUUUUUGUAAUUUAUAUUUAUUAC -------..(((.....((((.(((....))).)))).....)))....((((.(((....))))))).................(((((........))))) ( -14.50, z-score = -1.42, R) >droYak2.chr2L 9531594 89 - 22324452 ---------CGCCUACCUGCCUGAUGAAAGUCUGCCAUUAUUGUGAUUUAUGACAGAUUUUUGUUUAUCCAC-UU----UUUUGUGUAAUUUAUAUUUAUUAU ---------.............((..((((((((.(((...........))).))))))))..))....(((-..----....)))................. ( -12.80, z-score = -1.58, R) >droEre2.scaffold_4929 14307030 90 + 26641161 ---------CGCUUACCUGCCUGAUGAAAGUCUGCCAUUAAUGUGAUUUAUGACAGAUUUCUGUUUAUCCACUUU----UUUUGUGUCAUUUAUAUUUAUUAC ---------.((......))..((((((((((((.(((...........))).))))))).........(((...----....))))))))............ ( -11.40, z-score = -0.47, R) >droAna3.scaffold_12916 3843231 83 + 16180835 ---------CGCCGACCUGUCUGAUGAAAAUCUGUCAUUAAUGUGAUUUAUGACAGAUU--UCUGCACUU---------CCACUCGUAAUUUAUAUUUAUUAC ---------.((.((.(((((.(((....))).(((((....)))))....)))))...--)).))....---------......(((((........))))) ( -14.70, z-score = -1.85, R) >dp4.chr4_group4 814312 94 - 6586962 GGCCGGGAACCCGAGCCCGUCUGAUGAAAAUCUGUCAUUAAUGUGAUUUAUGACAGAUUUCUCUGCUCUU---------UCACUGGUAAUUUAUAUUUAUCAC ((((((....))).)))....(((((((((((((((((...........))))))))))....((((...---------.....)))).......))))))). ( -25.80, z-score = -2.88, R) >droPer1.super_46 343006 94 + 590583 GGCCGGGAACCCGAGCCCGUCUGAUGAAAAUCUGUCAUUAAUGUGAUUUAUGACAGAUUUCUCUGCUCUU---------UCACUGGUAAUUUAUAUUUAUCAC ((((((....))).)))....(((((((((((((((((...........))))))))))....((((...---------.....)))).......))))))). ( -25.80, z-score = -2.88, R) >droWil1.scaffold_180708 6183671 76 - 12563649 ---CGAGAAGGCAAA-----CUGAUGAAAAUCUGUCAUU---GUGAUUUAUGACAGAUU-------CUUU---------UCAGUUGUAAUUUAUAUUUAUUAC ---..(((..((((.-----((((.((.((((((((((.---.......))))))))))-------.)).---------))))))))..)))........... ( -20.80, z-score = -3.64, R) >consensus _________CGCUUACCUGCCUGAUGAAAGUCUGCCAUUAAUGUGAUUUAUGACAGAUUUCUCUUCAUCC_C_UU____UUUUUUGUAAUUUAUAUUUAUUAC ...........................(((((((.(((...........))).)))))))........................................... ( -6.14 = -6.11 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:56 2011