| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,230,489 – 1,230,589 |
| Length | 100 |
| Max. P | 0.630121 |

| Location | 1,230,489 – 1,230,589 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.54921 |
| G+C content | 0.61853 |
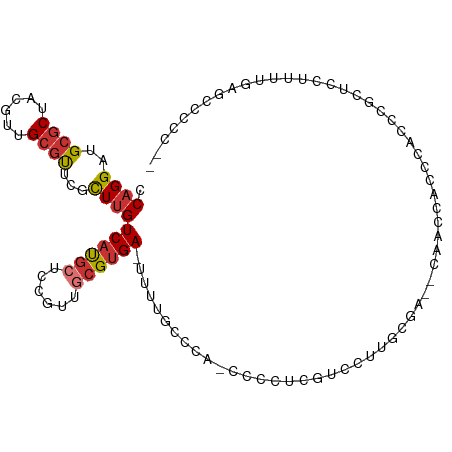
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -12.56 |
| Energy contribution | -13.11 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
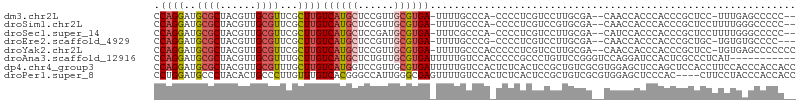
>dm3.chr2L 1230489 100 - 23011544 CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGUUGCGUGA-UUUUGCCCA-CCCCUCGUCCUUGCGA--CAACCACCCACCCGCUCC-UUUGAGCCCCC-- ...((.(((((.(((..(.((..((..(((((((......))))))-)...))..)-).)..)))....))).--)).))........((((.-...))))....-- ( -26.90, z-score = -2.72, R) >droSim1.chr2L 1202216 101 - 22036055 CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGUUGCGUGA-UUUUGCCCA-CCCCUCGUCCGUGCGA--CAACCACCCACCCGCUCCUUUUGGGCCCCC-- .((((..((((......))))...))))((((((......))))))-....(((((-......((..(((.(.--.......))))..))......)))))....-- ( -26.50, z-score = -0.82, R) >droSec1.super_14 1181336 101 - 2068291 CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGAUGCGUGA-UUUCGCCCA-CCCCUCGUCCUUGCGA--CAUCCACCCACCCGCUCCUUUUGGGCCCCC-- ...((((((((.(((..(.((..((..(((((((......))))))-)...))..)-).)..)))....))).--)))))........((.((....))))....-- ( -30.30, z-score = -2.55, R) >droEre2.scaffold_4929 1268533 99 - 26641161 CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGUUGCGUGA-UUUUGCCCG-CCCCUCGUCCUUGCGA--CAACCACCCACCCGCUGC-UGUGUGCCCC--- ...((.(((((.(((..(((...((..(((((((......))))))-)...)).))-)....)))....))).--)).)).......(((...-.)))......--- ( -25.40, z-score = -0.67, R) >droYak2.chr2L 1205133 103 - 22324452 CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGUUGCGUGA-UUUUGCCCACCCCCUCGUCCUUGCGA--CAACCACCCACCCGCUCC-UGUGAGCCCCCCC ..(((((((((......))))..((..(((((((......))))))-)...))..........))))).....--.............((((.-...))))...... ( -25.50, z-score = -1.91, R) >droAna3.scaffold_12916 4691937 96 - 16180835 CCAGGAUGCGCUACGUUGCGUUUGCUUGUCAUGCUCUGUUGCGUGAUUUUUGUCCACCCCCGCCCUGUUCCGGGUCCAGGAUCCACUCGCCCUCAU----------- ...((((((((......))))......(((((((......)))))))....)))).((...((((......))))...))................----------- ( -25.30, z-score = -0.45, R) >dp4.chr4_group3 9682357 107 - 11692001 CCAGGAUGCGCUACGUUGCGUUUGCUUGUCAUGGUCCGUUGCGUGAUUUUUGUCCACUCUCACUCCGCUGUCGCGUGGAGCUCCAGCUCCACCUUCCACCCACCACC .((((..((((......))))...))))...((((..((...(((((....((........))......)))))(((((((....))))))).....))..)))).. ( -28.90, z-score = -0.77, R) >droPer1.super_8 880753 103 - 3966273 CCUGGAUGCCCUACACUGCCCUUGUUUGUCACGGGCCAUUGGGCGAGUUUUGUCCACUCUCACUCCGCUGUCGCGUGGAGCUCCCAC----CUUCCUACCCACCACC ...(((.((((......((((.((.....)).))))....))))((((.......))))...((((((......)))))).......----..)))........... ( -29.00, z-score = -0.85, R) >consensus CCAGGAUGCGCUACGUUGCGUUCGCUUGUCAUGCUCCGUUGCGUGA_UUUUGCCCA_CCCCUCGUCCUUGCGA__CAACCACCCACCCGCUCCUUUUGAGCCCCC__ .((((..((((......))))...))))((((((......))))))............................................................. (-12.56 = -13.11 + 0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:57 2011