| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,109,198 – 13,109,315 |
| Length | 117 |
| Max. P | 0.971358 |

| Location | 13,109,198 – 13,109,315 |
|---|---|
| Length | 117 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.63268 |
| G+C content | 0.53625 |
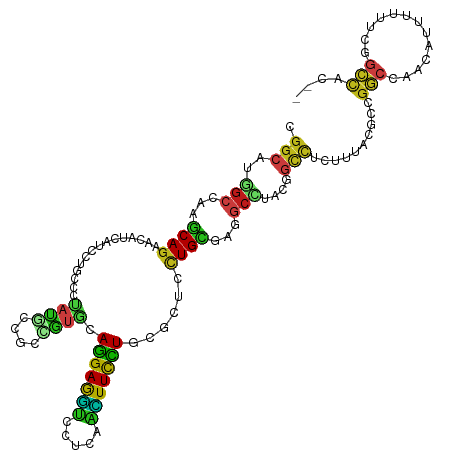
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -18.17 |
| Energy contribution | -16.89 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
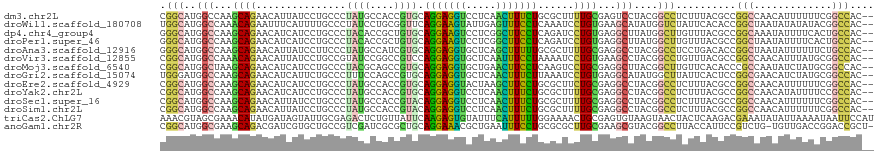
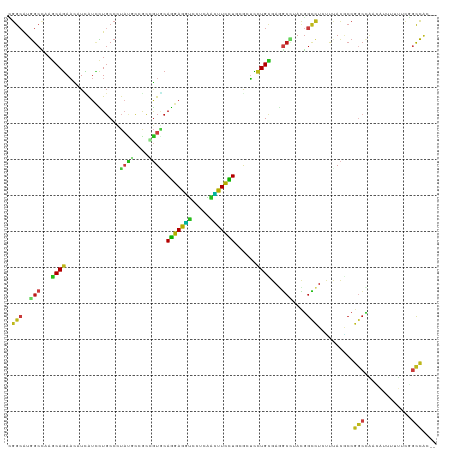
>dm3.chr2L 13109198 117 + 23011544 CGGCAUGGCCAAGCAGAACAUUAUCCUGCCCUAUGCCACCGUGCAGGAGGUCCUCAACUUUCUGCGCUUUUGCGAGUCCUACGGCCUCUUUACGCCGGCCAACAUUUUUUCGGCCAC-- .(((((((....((((.........)))).)))))))...(((((((((((.....)))))))))))...............(((........)))((((...........))))..-- ( -40.60, z-score = -2.69, R) >droWil1.scaffold_180708 6182825 117 + 12563649 UGGCAUGGCCAAACAGAAUUUCAUUUUGCCCUAUCCUGCGGUUCAGGAAGUAUUGAGUUUCCUCAAAUCCUGUGAAGCAUAUGGUCUAUUCACACCGGCUAAUAUAUAUACGGCCAC-- .....(((((.....(.....).....(((......(((....(((((....(((((....))))).)))))....)))...(((........))))))............))))).-- ( -27.60, z-score = -0.35, R) >dp4.chr4_group4 813644 117 + 6586962 GGGCAUGGCCAAGCAGAACAUCAUCCUGCCCUACACCGCUGUGCAGGAAGUCCUCGGCUUCCUCAGAUCCUGUGAGGCUUAUGGCUUGUUUACGCCGGCAAAUAUUUUCACUGCCAC-- .(((.((..(((((.(.....).....(((.((((...(((...(((((((.....))))))))))....)))).))).....)))))..)).)))((((...........))))..-- ( -36.20, z-score = -0.49, R) >droPer1.super_46 342338 117 - 590583 GGGCAUGGCCAAGCAGAACAUCAUCCUGCCCUACACCGCUGUGCAGGAAGUCCUCGGCUUCCUCAGAUCCUGUGAGGCUUAUGGCUUGUUUACGCCGGCUAAUAUUUUCACUGCCAC-- .(((((((((..((.(((((((((...(((.((((...(((...(((((((.....))))))))))....)))).)))..))))..)))))..)).)))))..........))))..-- ( -36.10, z-score = -0.46, R) >droAna3.scaffold_12916 3842779 117 - 16180835 GGGCAUGGCCAAGCAGAACAUUAUCCUUCCCUAUGCCAUCGUGCAGGAGGUGCUCAGCUUUUUGCGCUUUUGCGAGGCCUACGGCCUCCUGACACCGGCUAAUAUUUUUUCUGCCAC-- .(((((((((..(((((((((...........))).....(((((((((((.....)))))))))))))))))((((((...))))))........)))))..........))))..-- ( -39.40, z-score = -1.43, R) >droVir3.scaffold_12855 2320262 117 - 10161210 CGGCAUGGCCAAACAGAACAUUAUCCUGCCGUAUCCGGCCGUCCAGGAGGUGCUCAAUUUCCUAAAAUCCUGUGAAGCCUACGGCCUGUUUACGCCGGCCAACAUUUAUGCGGCCAC-- .(((..(((...((((...........((((....)))).....(((((((.....)))))))......))))...)))...((((.((....)).))))............)))..-- ( -35.80, z-score = -0.84, R) >droMoj3.scaffold_6540 11086106 117 - 34148556 CGGCAUGGCUAAGCAGAACAUCAUCCUGCCCUACGCAGCCGUGCAGGAGGUGCUGAACUUCCUCAAGUCCUGCGAGGCUUACGGCUUGUUCACACCCGCCAAUAUCUAUGCGGCCAC-- .(((........((((.........))))....(((((((.(((((((..((..(.....)..))..))))))).)))....(((..((....))..)))........)))))))..-- ( -37.50, z-score = -0.31, R) >droGri2.scaffold_15074 3333991 117 - 7742996 UGGGAUGGCCAAGCAGAACAUCAUUCUGCCCUUUCCAGCCGUGCAGGAGGUGCUCAACUUUCUUAAAUCCUGUGAGGCAUAUGGCUUAUUCACUCCGGCGAACAUCUAUGCGGCCAC-- .....(((((..((((((.....))))))........((((...(((((((.....)))))))........(((((((.....)))...))))..))))............))))).-- ( -34.00, z-score = 0.01, R) >droEre2.scaffold_4929 14306476 117 - 26641161 CGGCAUGGCCAAGCAGAACAUCAUCCUGCCCUAUGCCACCGUGCAGGAGGUACUAAGCUUCCUGCGCUUCUGCGAGGCCUACGGCCUCUUUACGCCGGCCAACAUUUUUUCGGCCAC-- .(((((((....((((.........)))).)))))))...(((((((((((.....)))))))))))......((((((...))))))........((((...........))))..-- ( -50.40, z-score = -4.35, R) >droYak2.chr2L 9531020 117 + 22324452 CGGCAUGGCCAAGCAGAACAUCAUCCUGCCCUAUGCCACCGUGCAGGAGGUCCUCAACUUUCUGCGCUUUUGCGAGGCCUACGGCCUCUUUACGCCGGCCAACAUAUUUUCCGCCAC-- .(((((((....((((.........)))).)))))))...(((((((((((.....)))))))))))......((((((...))))))........(((.............)))..-- ( -42.82, z-score = -3.12, R) >droSec1.super_16 1264409 117 + 1878335 CGGCAUGGCCAAGCAGAACAUUAUCCUGCCCUAUGCCACCGUACAGGAGGUCCUCAACUUUCUGCGCUUUUGCGAGGCCUACGGCCUCUUUACGCCGGCCAACAUUUUUUCGGCCAC-- .(((((((....((((.........)))).)))))))..((((((((((((.....)))))))).((....))((((((...))))))..))))..((((...........))))..-- ( -42.60, z-score = -3.28, R) >droSim1.chr2L 12901388 117 + 22036055 CGGCAUGGCCAAGCAGAACAUUAUCCUGCCCUAUGCCACCGUACAGGAGGUCCUCAACUUUCUGCGCUUUUGCGAGGCCUACGGCCUCUUUACGCCGGCCAACAUUUUUUCGGCCAC-- .(((((((....((((.........)))).)))))))..((((((((((((.....)))))))).((....))((((((...))))))..))))..((((...........))))..-- ( -42.60, z-score = -3.28, R) >triCas2.ChLG7 8379722 119 - 17478683 AAACGUAGCGAAACAUAUGAUAGUAUUGCGAGACUCUGUUAUUCAAGAGUGUAUUUCAUUUUUGGAAAACUGCGAGUGUAAGUAACUACUCAAGACGAAAUAUAUUAAAAUAAUUCCAU ...(((.((((.((........)).))))(((((((.(((.((((((((((.....)))))))))).)))...))))((.....))..)))...)))...................... ( -22.80, z-score = -0.99, R) >anoGam1.chr2R 28575096 117 + 62725911 CGGCAUGGCGAAGCAGACGAUCGUGCUGCCGUCGAUCGCGCUGCAGGAAACGCUGAAUUUCCUGCGCGCUUGCGAAGCGUACGGCCUUACCAUUCCGUCUG-UGUUGACCGGACCGCU- (((..((((((.(((((((...((((.((..(((...((((.((((((((.......))))))))))))...))).))))))((.....))....))))))-).))).)))..)))..- ( -50.40, z-score = -2.04, R) >consensus CGGCAUGGCCAAGCAGAACAUCAUCCUGCCCUAUGCCGCCGUGCAGGAGGUCCUCAACUUCCUGCGCUCCUGCGAGGCCUACGGCCUCUUUACGCCGGCCAACAUUUUUUCGGCCAC__ .(((..(((...((((...............((((....)))).(((((((.....)))))))......))))...)))....)))..........(((.............))).... (-18.17 = -16.89 + -1.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:55 2011