
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,081,501 – 13,081,606 |
| Length | 105 |
| Max. P | 0.541985 |

| Location | 13,081,501 – 13,081,606 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Shannon entropy | 0.49455 |
| G+C content | 0.32321 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -9.93 |
| Energy contribution | -9.64 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541985 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
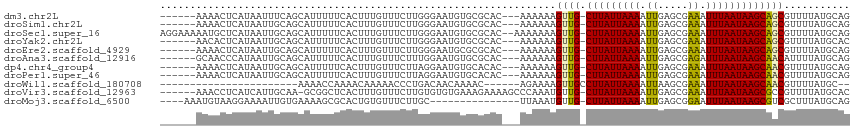
>dm3.chr2L 13081501 105 - 23011544 ------AAAACUCAUAAUUUCAGCAUUUUUCACUUUGUUUCUUGGGAAUGUGCGCAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAG ------................((((((..((..........))..)).))))(((.---((((..((((-(((((((((.((.....)).))))))))))))).)))).))).. ( -24.10, z-score = -1.60, R) >droSim1.chr2L 12871708 105 - 22036055 ------AAAACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUGGGAAUGUGCGCAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAG ------....(((((((.(((.((((((..((..........))..)).))))))).---......((((-(((((((((.((.....)).)))))))))))))...))))).)) ( -24.70, z-score = -1.20, R) >droSec1.super_16 1236598 112 - 1878335 AGGAAAAAUGCUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUGGGAAUGUGCGCAC--AAAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAG ..((((((((((((....)).)))))))))).....((((.....))))....(((.--((((...((((-(((((((((.((.....)).))))))))))))).)))).))).. ( -32.30, z-score = -2.76, R) >droYak2.chr2L 9501756 105 - 22324452 ------AACACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUGGGAAUGUGCGCAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAC ------................((((((..((..........))..)).))))(((.---((((..((((-(((((((((.((.....)).))))))))))))).)))).))).. ( -24.10, z-score = -1.09, R) >droEre2.scaffold_4929 14279003 105 + 26641161 ------AAAACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUGGGAAUGCGCGCAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAG ------.............((.((((((((.............))))))))))(((.---((((..((((-(((((((((.((.....)).))))))))))))).)))).))).. ( -25.02, z-score = -1.18, R) >droAna3.scaffold_12916 3815307 105 + 16180835 ------GCAACCCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUUGGAAUGUGCGCAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAGAUUUAAUAAGCAACAUUUUAUGCAG ------((((.......)))).((((.(((((...........))))).))))(((.---.((((.((((-(((((((((.((....))..))))))))))))).)))).))).. ( -26.70, z-score = -2.29, R) >dp4.chr4_group4 778361 105 - 6586962 ------AAAACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUAGGAAUGUGCACAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAACGUUUUAUGCAG ------............((((((((.((((.............)))).))))....---.((((.((((-(((((((((.((.....)).))))))))))))).)))).)))). ( -24.02, z-score = -1.87, R) >droPer1.super_46 307233 105 + 590583 ------AAAACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUAGGAAUGUGCACAC---AAAAAAGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAACGUUUUAUGCAG ------............((((((((.((((.............)))).))))....---.((((.((((-(((((((((.((.....)).))))))))))))).)))).)))). ( -24.02, z-score = -1.87, R) >droWil1.scaffold_180708 6139965 84 - 12563649 -----------------------AAAACCAAAACAAAAACCCUGACAACAAAAC------AGAAAAGUUGCCUUAUUAAAAUUAAGCGAAAUUUAAUAAGCAACGUUUUAUGC-- -----------------------......(((((.......(((.........)------))....(((((.((((((((.((.....)).))))))))))))))))))....-- ( -13.40, z-score = -3.24, R) >droVir3.scaffold_12963 6949343 107 - 20206255 ------AAACCUCAUCAUUGCAA-GCGGCUCACUUUGUUUCUUGUGUGUGAAAGAAAAGCCCAAAUGUUG-CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCGCCGUUUUAUGCAC ------............(((((-(((((........((((((........))))))............(-(((((((((.((.....)).))))))))))))))))...)))). ( -25.00, z-score = -1.43, R) >droMoj3.scaffold_6500 26137832 95 - 32352404 ----AAAUGUAAGGAAAAUUGUGAAAAGCGCACUGUGUUUCUUGC---------------UUAAAUGUUG-CUUAUUAAAAUUGAGCGGAAUUUAAUAAGCGUCGCUUUAUGCAG ----....((((((((....(((.......)))....))))))))---------------((((((.(((-((((.......))))))).))))))...((((......)))).. ( -21.10, z-score = -0.80, R) >consensus ______AAAACUCAUAAUUGCAGCAUUUUUCACUUUGUUUCUUGGGAAUGUGCGCAC___AAAAAAGUUG_CUUAUUAAAAUUGAGCGAAAUUUAAUAAGCAGCGUUUUAUGCAG ..................................................................((((.(((((((((.((.....)).)))))))))))))........... ( -9.93 = -9.64 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:53 2011