
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,076,864 – 13,076,970 |
| Length | 106 |
| Max. P | 0.878478 |

| Location | 13,076,864 – 13,076,970 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 69.94 |
| Shannon entropy | 0.61432 |
| G+C content | 0.45361 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -10.79 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
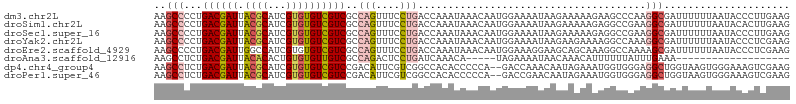
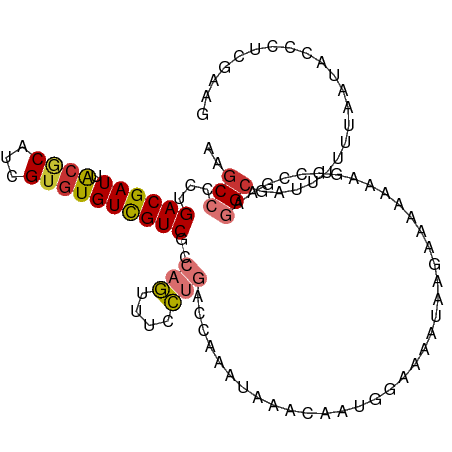
>dm3.chr2L 13076864 106 + 23011544 AAGCCCCUGACGAUUACGCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAAUAAGAAAAAGAAGCCCAAGGCGAUUUUUUAAUACCCUUGAAG ..((..(((.((((.((((.....)))).)))).)))(((((................))))).............)).(((((..(((....)))..)))))... ( -20.49, z-score = -1.40, R) >droSim1.chr2L 12867162 106 + 22036055 AAGCCCCUGACGAUUACGCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAAUAAGAAAAAGAGGCCGAAGGCGAUUUUUUAAUACACUUGAAG ..(((...((((((.((((...))))))))))(((..(((((................)))))............)))....)))..................... ( -19.93, z-score = -0.14, R) >droSec1.super_16 1231970 106 + 1878335 AAGCCCCUGACGAUUACGCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAAUAAGAAAAAGAGGCCGAAGGCGAUUUUUUAAUACCCUUGAAG ..(((.(((.((((.((((.....)))).)))).)))(((((................)))))............)))..((((..(((....)))..)))).... ( -20.89, z-score = -0.59, R) >droYak2.chr2L 9496925 106 + 22324452 AAGCCCCUGACGAUUACGCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAAUAAGAAGAAAAGGCCAAAGGCGAUUUUUUAAUACCCUCGAAG ..(((...((((((.((((...))))))))))(((..(((((................)))))............)))....)))..................... ( -19.93, z-score = -0.40, R) >droEre2.scaffold_4929 14274535 106 - 26641161 AAGCCCCUGACGAUUGGCCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAGGAAGCAGCAAAGGCCAAAAGCGAUUUUUUAAUACCCUCGAAG ..(((...((((((..((....))..))))))((..(((((((..(((.........)))..))))))).))...)))............................ ( -25.40, z-score = -1.08, R) >droAna3.scaffold_12916 3811439 82 - 16180835 AAGCCUCUGACGAUUACACACUGUGUGUUGUCGCCAGACUCCUGAUCAAACA-----UAGAAAAUAACAAACAUUUUUUAUUUGAAA------------------- .....((((.((((.((((.....)))).)))).)))).......((((..(-----((((((((.......)))))))))))))..------------------- ( -18.20, z-score = -2.98, R) >dp4.chr4_group4 772830 104 + 6586962 AAGCCUCUGACGAUUACGCAUCGUGUGUCGUCCGACAUUCGUCGGCCACACCCCCA--GACCAAACAAUAGAAAUGGUGGGAGGCUGGUAAGUGGGAAAGUCGAAG ...((((.((((((.((((...)))))))))).))((((..((((((...((((((--...(........)...))).))).))))))..)))))).......... ( -32.50, z-score = -0.83, R) >droPer1.super_46 301672 104 - 590583 AAGCCUCUGACGAUUACGCAUCGUGUGUCGUCCGACAUUCGUCGGCCACACCCCCA--GACCGAACAAUAGAAAUGGUGGGAGGCUGGUAAGUGGGAAAGUCGAAG ...((((.((((((.((((...)))))))))).))((((..((((((...((((((--...(........)...))).))).))))))..)))))).......... ( -33.20, z-score = -0.74, R) >consensus AAGCCCCUGACGAUUACGCAUCGUGUGUCGUCGCCAGUUUCCUGACCAAAUAAACAAUGGAAAAUAAGAAAAAAAGGCCGAAGGCGAUUUUUUAAUACCCUCGAAG ..(((...((((((.((((...))))))))))..(((....)))......................................)))..................... (-10.79 = -11.22 + 0.44)
| Location | 13,076,864 – 13,076,970 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Shannon entropy | 0.61432 |
| G+C content | 0.45361 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -11.74 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
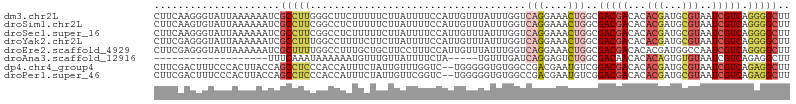
>dm3.chr2L 13076864 106 - 23011544 CUUCAAGGGUAUUAAAAAAUCGCCUUGGGCUUCUUUUUCUUAUUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGCGUAAUCGUCAGGGGCUU ...((((((.(((....)))).)))))(((((((..........................(((((....)))))(((((...(((...)))..)))))))))))). ( -28.10, z-score = -1.91, R) >droSim1.chr2L 12867162 106 - 22036055 CUUCAAGUGUAUUAAAAAAUCGCCUUCGGCCUCUUUUUCUUAUUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGCGUAAUCGUCAGGGGCUU ......(((.(((....))))))....(((((((..........................(((((....)))))(((((...(((...)))..)))))))))))). ( -26.40, z-score = -1.49, R) >droSec1.super_16 1231970 106 - 1878335 CUUCAAGGGUAUUAAAAAAUCGCCUUCGGCCUCUUUUUCUUAUUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGCGUAAUCGUCAGGGGCUU .....((((((((....))).))))).(((((((..........................(((((....)))))(((((...(((...)))..)))))))))))). ( -28.50, z-score = -1.92, R) >droYak2.chr2L 9496925 106 - 22324452 CUUCGAGGGUAUUAAAAAAUCGCCUUUGGCCUUUUCUUCUUAUUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGCGUAAUCGUCAGGGGCUU ...((((((((((....))).)))))))((((((..........................(((((....)))))(((((...(((...)))..))))))))))).. ( -29.40, z-score = -1.83, R) >droEre2.scaffold_4929 14274535 106 + 26641161 CUUCGAGGGUAUUAAAAAAUCGCUUUUGGCCUUUGCUGCUUCCUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGGCCAAUCGUCAGGGGCUU ...((((((((((....))).)))))))(((((((.((........(((((((.....(.(((((....))))).)......))))))).....)).))))))).. ( -29.02, z-score = -0.69, R) >droAna3.scaffold_12916 3811439 82 + 16180835 -------------------UUUCAAAUAAAAAAUGUUUGUUAUUUUCUA-----UGUUUGAUCAGGAGUCUGGCGACAACACACAGUGUGUAAUCGUCAGAGGCUU -------------------..(((((((.((((((.....))))))...-----)))))))..(((..((((((((..((((.....))))..))))))))..))) ( -23.30, z-score = -3.19, R) >dp4.chr4_group4 772830 104 - 6586962 CUUCGACUUUCCCACUUACCAGCCUCCCACCAUUUCUAUUGUUUGGUC--UGGGGGUGUGGCCGACGAAUGUCGGACGACACACGAUGCGUAAUCGUCAGAGGCUU ....................((((((((.(((..((........))..--)))))((((.((((((....))))).).))))(((((.....)))))..)))))). ( -33.90, z-score = -1.27, R) >droPer1.super_46 301672 104 + 590583 CUUCGACUUUCCCACUUACCAGCCUCCCACCAUUUCUAUUGUUCGGUC--UGGGGGUGUGGCCGACGAAUGUCGGACGACACACGAUGCGUAAUCGUCAGAGGCUU ....................((((((((.(((..((........))..--)))))((((.((((((....))))).).))))(((((.....)))))..)))))). ( -33.30, z-score = -0.94, R) >consensus CUUCGAGGGUAUUAAAAAAUCGCCUUCGGCCUUUUCUUCUUAUUUUCCAUUGUUUAUUUGGUCAGGAAACUGGCGACGACACACGAUGCGUAAUCGUCAGGGGCUU .....................(((((....................................(((....)))..(((((...(((...)))..))))).))))).. (-11.74 = -12.27 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:50 2011