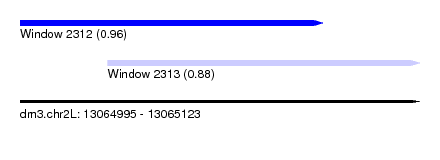
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,064,995 – 13,065,123 |
| Length | 128 |
| Max. P | 0.956056 |

| Location | 13,064,995 – 13,065,092 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.32 |
| Shannon entropy | 0.63750 |
| G+C content | 0.52320 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.79 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.956056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
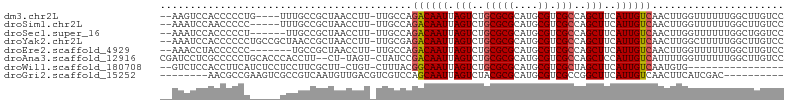
>dm3.chr2L 13064995 97 + 23011544 --AAGUCCACCCCCUG----UUUGCCGCUAACCUU-UUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCC --..............----...((((..((((..-......((((((.(((..(((((....)).)))..)))..)))))).....))))...))))...... ( -22.22, z-score = -0.63, R) >droSim1.chr2L 12855274 96 + 22036055 --AAAUCCAACCCCC-----UUUGCCGCUAACCUU-UUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCC --.............-----...((((..((((..-......((((((.(((..(((((....)).)))..)))..)))))).....))))...))))...... ( -22.22, z-score = -1.26, R) >droSec1.super_16 1220509 95 + 1878335 --AAAUCCACCCCCU------UUGCCGCUAACCUU-UUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUGGUCC --..........((.------..((((..((((..-......((((((.(((..(((((....)).)))..)))..)))))).....))))...)))).))... ( -23.92, z-score = -1.21, R) >droYak2.chr2L 9484987 101 + 22324452 --AAAUCCACCCCCCUGCCGCUAACCGCUAACCUU-UUGCGAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGCUUUUUGGCUUGUCC --..............((((((((.(((.......-..))).((((((.(((..(((((....)).)))..)))..))))))...))))).....)))...... ( -24.60, z-score = -1.10, R) >droEre2.scaffold_4929 14261740 94 - 26641161 --AAACCUACCCCCC-------UGCCGCUAACCUU-UUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCC --.............-------.((((..((((..-......((((((.(((..(((((....)).)))..)))..)))))).....))))...))))...... ( -22.22, z-score = -1.30, R) >droAna3.scaffold_12916 3800479 100 - 16180835 CGAUCCUCGCCCCCUGCACCCACCUU--CU-UAGU-CUAUCCGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUCCAUUGUCAUUUUGGUUUUUUGGCUUGUCC ........(((......(((...((.--..-.)).-......((((((.(((..(((((....)).)))..)))..)))))).....))).....)))...... ( -18.90, z-score = -0.37, R) >droWil1.scaffold_180708 6112361 84 + 12563649 --GUCUCCACCUUCAUCUCCUCCUUCGCUU-CUGU-CUUUACGGCAAUUAGUCUGCGCGCAUGCGUCGCUAGCUUCAUUGUCAAUGUG---------------- --............................-....-......((((((.(((..(((((....)).)))..)))..))))))......---------------- ( -15.40, z-score = -1.57, R) >droGri2.scaffold_15252 7093793 86 - 17193109 --------AACGCCGAAGUCGCCGUCAAUGUUGACGUCGUCCAGCAAUUAGUCUACGCGCAUGCGUCGCCGGCUUCAUUGUCAACUUCAUCGAC---------- --------..((..(((((((.(((((....))))).))....(((((.((((..((((....)).))..))))..)))))..)))))..))..---------- ( -21.30, z-score = 0.17, R) >consensus __AAAUCCACCCCCC_____UUUGCCGCUAACCUU_UUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCC ..........................................((((((.(((..(((((....)).)))..)))..))))))...................... (-13.73 = -13.79 + 0.06)

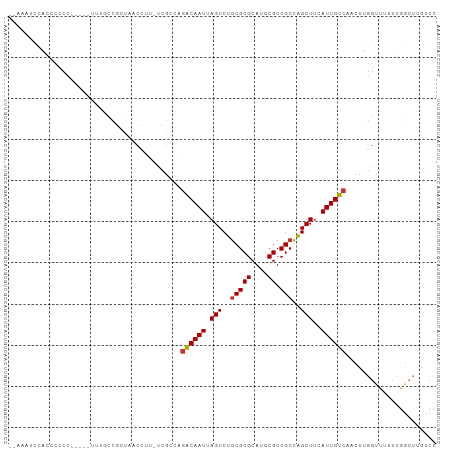
| Location | 13,065,023 – 13,065,123 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.32 |
| Shannon entropy | 0.15811 |
| G+C content | 0.54042 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
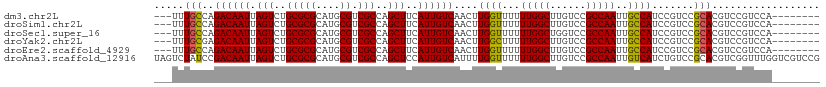
>dm3.chr2L 13065023 100 + 23011544 ---UUUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA-------- ---..(((..((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..)))).......)))..........-------- ( -28.80, z-score = -1.82, R) >droSim1.chr2L 12855301 100 + 22036055 ---UUUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA-------- ---..(((..((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..)))).......)))..........-------- ( -28.80, z-score = -1.82, R) >droSec1.super_16 1220535 100 + 1878335 ---UUUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUGGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA-------- ---..(((..((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..)))).......)))..........-------- ( -28.80, z-score = -1.16, R) >droYak2.chr2L 9485019 100 + 22324452 ---UUUGCGAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGCUUUUUGGCUUGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA-------- ---..((((.((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..))))......))))..........-------- ( -33.10, z-score = -2.43, R) >droEre2.scaffold_4929 14261765 100 - 26641161 ---UUUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA-------- ---..(((..((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..)))).......)))..........-------- ( -28.80, z-score = -1.82, R) >droAna3.scaffold_12916 3800507 111 - 16180835 UAGUCUAUCCGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUCCAUUGUCAUUUUGGUUUUUUGGCUUGUCCGCCAAUUGUCAUCUGUCCGCACGUCGGUUUGGUCGUCCG ..(.(((.(((((.........(((((....)).)))..((.(((.........)))....(((((......))))).............))..)))))..))).)..... ( -25.80, z-score = 0.00, R) >consensus ___UUUGCCAGACAAUUAGUCUGCGCGCAUGCGUCGCCAGCUUCAUUGUCAACUUGGUUUUUUGGCUUGUCCGCCAAUUGCCAUCCGUCCGCACGUCCGUCCA________ .....(((..((((((.(((..(((((....)).)))..)))..))))))....((((...(((((......)))))..)))).......))).................. (-27.33 = -27.08 + -0.25)
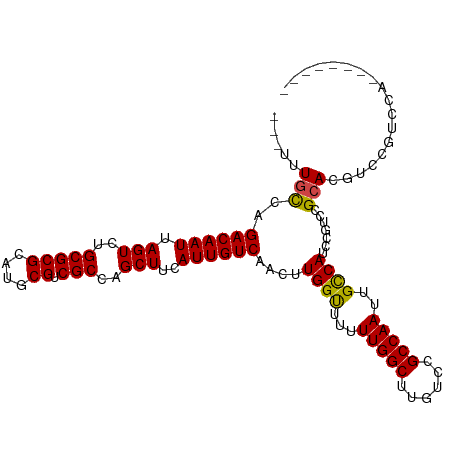
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:48 2011