
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,062,554 – 13,062,663 |
| Length | 109 |
| Max. P | 0.525370 |

| Location | 13,062,554 – 13,062,663 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Shannon entropy | 0.33640 |
| G+C content | 0.41329 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525370 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 13062554 109 - 23011544 -----------UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGCGCAGAGUUUU -----------...........(((((...((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))))))).. ( -28.60, z-score = -0.75, R) >droSim1.chr2L 12852799 109 - 22036055 -----------UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGCGCAGAGUUUG -----------...........(((((...((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))))))).. ( -28.60, z-score = -0.72, R) >droSec1.super_16 1218078 109 - 1878335 -----------UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGCGCAGAGUUUG -----------...........(((((...((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))))))).. ( -28.60, z-score = -0.72, R) >droYak2.chr2L 9482505 109 - 22324452 -----------UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGUGCAGAGUGUU -----------.(((.......))).....((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))....... ( -26.10, z-score = 0.21, R) >droEre2.scaffold_4929 14259087 109 + 26641161 -----------UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGCGCAGAGUUUU -----------...........(((((...((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))))))).. ( -28.60, z-score = -0.75, R) >droAna3.scaffold_12916 3798144 103 + 16180835 -----------------CCUUCAACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCAUUUCUUUAGCUGCUUUUAGCGCAGACUUUU -----------------.............((((((..((((.(((....(((..(((((....)))))..)))....(((....))).........)))))))...))))))....... ( -26.60, z-score = -1.73, R) >dp4.chr4_group4 754026 115 - 6586962 -----UCGCGCUGUCACCUCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCCGCUUUUAGCAGAGACUCUU -----..((((.(((.......)))......))))..((((.((((....)))).(((((....)))))..))))...((((.(((.......))).))))((.....)).......... ( -26.50, z-score = 0.51, R) >droWil1.scaffold_180708 6107870 117 - 12563649 AAAAAUGAAGAAAGAAAUACACAGACUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGUUUAAGCUUAUUUCUUUGGCUGCUUUUAGCAAAAUUU--- ..........(((((((((...((.(((((...(((.((((.((((....)))).(((((....)))))..))))...))))))))))))))))))).((((....)))).......--- ( -25.00, z-score = -0.56, R) >droVir3.scaffold_12963 6925154 92 - 20206255 ----------------------AAACUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUGAUUUC-UUGGCCGCUUUUAGCGCAUU----- ----------------------......((((((((..(((.((((....)))).(((((....)))))..((((.(..((....))..)....-.)))).)))...))))))))----- ( -28.70, z-score = -2.29, R) >droMoj3.scaffold_6500 26108984 92 - 32352404 ----------------------AAACUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUCGCUUAAGCUGAUUUC-UUGGCCGCUUUUAGCGCAUU----- ----------------------......((((((((..(((.((((....)))).(((((....)))))..((((..((((....)).))....-.)))).)))...))))))))----- ( -26.50, z-score = -1.85, R) >droGri2.scaffold_15252 10094384 92 - 17193109 ----------------------AAACUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUGAUUUC-UUGGCCGCUUUUAGCGCAUU----- ----------------------......((((((((..(((.((((....)))).(((((....)))))..((((.(..((....))..)....-.)))).)))...))))))))----- ( -28.70, z-score = -2.29, R) >consensus ___________UGUCACCCCUCGACUUUAAUGCGCUUUGGCAUGCUUUUAGGCAAUAAUUCAUGGAUUAACGCCAUUUGGCUUAAGCUCGUUUCUUUGGCUGCUUUUAGCGCAGAGUUUU ..............................((((((..(((..(((....(((..(((((....)))))..)))....)))....(((.........))).)))...))))))....... (-22.14 = -22.35 + 0.21)

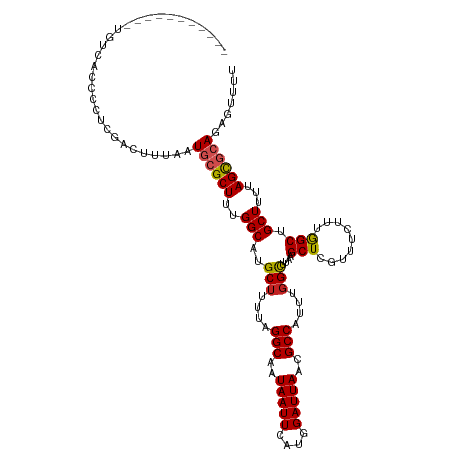
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:47 2011