| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,056,068 – 13,056,193 |
| Length | 125 |
| Max. P | 0.514380 |

| Location | 13,056,068 – 13,056,193 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Shannon entropy | 0.44491 |
| G+C content | 0.35084 |
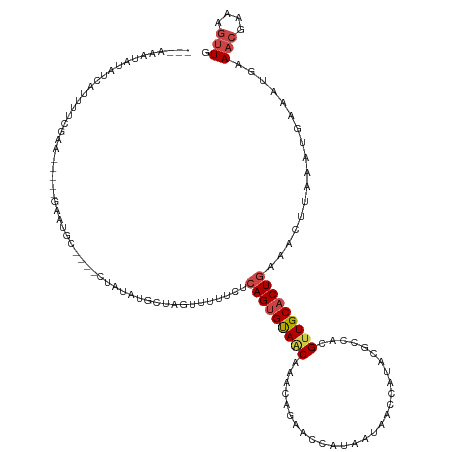
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
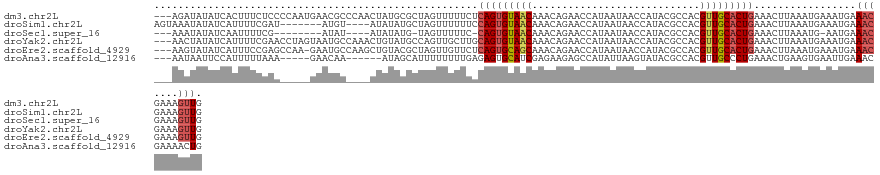
>dm3.chr2L 13056068 125 - 23011544 ---AGAUAUAUCACUUUCUCCCCAAUGAACGCCCAACUAUGCGCUAGUUUUUCUCAGUGUAACAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUGAAAUGAAACGAAAGUUG ---.......(((.((((...........(((........)))..........((((((((((............................)))))))))).........)))))))(((....))). ( -20.09, z-score = -1.86, R) >droSim1.chr2L 12846284 117 - 22036055 AGUAAAUAUAUCAUUUUCGAU-------AUGU----AUAUAUGCUAGUUUUUUCCAGUGUAACAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUGAAAUGAAACGAAAGUUG .....(((((((......)))-------))))----...(((...(((((....(((((((((............................))))))))))))))...)))......(((....))). ( -20.19, z-score = -1.57, R) >droSec1.super_16 1211607 110 - 1878335 ---AAAUAUAUCAAUUUUCG--------AUAU----AUAUAUG-UAGUUUUUC-CAGUGUAACAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUG-AAUGAAACGAAAGUUG ---........(((((((((--------...(----((.(((.-.(((((...-(((((((((............................))))))))))))))...)))-.)))...))))))))) ( -20.99, z-score = -2.85, R) >droYak2.chr2L 9475791 125 - 22324452 ---AACUAUAUCAUUUUCGAACCUAGUAAUGCCAAACUGUAUGCCAGUUGCUUGCAGUGUAACAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUGAAAUGAAACGAAAGUUG ---.......(((((((.......(((...((..(((((.....)))))))...(((((((((............................)))))))))..))).....)))))))(((....))). ( -22.59, z-score = -1.45, R) >droEre2.scaffold_4929 14252298 124 + 26641161 ---AAGUAUAUCAUUUCCGAGCCAA-GAAUGCCAAGCUGUACGCUAGUUGUUCUCAGUGCAGCAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUGAAAUGAAACGAAAGUUG ---.......(((((((........-((((..(.(((.....))).)..))))((((((((((............................)))))))))).........)))))))(((....))). ( -29.49, z-score = -2.69, R) >droAna3.scaffold_12916 3791615 114 + 16180835 ---AAUAAUUCCAUUUUUAAA-----GAACAA------AUAGCAUUUUUUUUGAGAGUGCAUCGAGAAGAGCCAUAUUAAGUAUACGCCACGUUGCCCUGAAACUGAAGUGAAUUGAAACGAAAACUG ---..((((((.(((((...(-----(..(((------...((.((((((((((.......)))))))))).........(....)))....)))..))......)))))))))))............ ( -12.70, z-score = 1.04, R) >consensus ___AAAUAUAUCAUUUUCGAA_____GAAUGC____CUAUAUGCUAGUUUUUCUCAGUGUAACAAACAGAACCAUAAUAACCAUACGCCACGUUGCACUGAAACUUAAAUGAAAUGAAACGAAAGUUG ......................................................(((((((((............................))))))))).................(((....))). (-10.42 = -10.92 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:45 2011