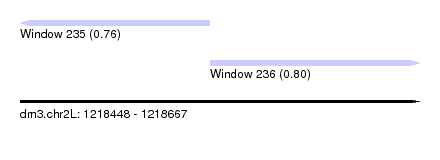
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,218,448 – 1,218,667 |
| Length | 219 |
| Max. P | 0.802743 |

| Location | 1,218,448 – 1,218,552 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.08 |
| Shannon entropy | 0.25737 |
| G+C content | 0.35669 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
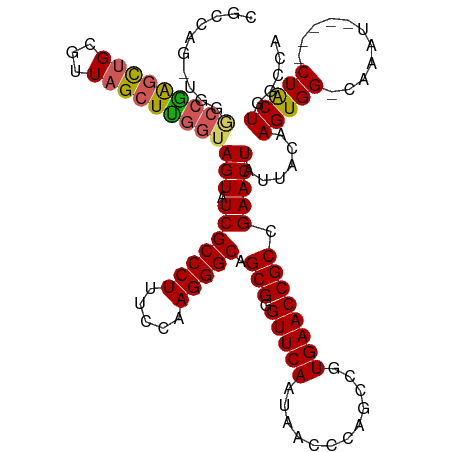
>dm3.chr2L 1218448 104 - 23011544 UCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAUGUAUUGCUAGUCAGUCAAAAAAGUGUGGGUAUUCCCAGGCGUGAAUAA---------------- .........(..(((((((..((((((((........))))))))............))))))...............((.((((....)))).)))..)....---------------- ( -24.04, z-score = -2.08, R) >droEre2.scaffold_4929 1264010 120 - 26641161 UCGUUACUAUUGCGCAACCAUAGCCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAUGUAUUGCCAGUCAGAGCAAAAAGUGUGGGUAUUCCCAAGCGUGAAUUGUGAAUCCGAUAAAUAC (((((((..(..(((.......(((((((........))))(((((((.((.....)).)))..)))).).))........((((....)))).)))..)...))))...)))....... ( -24.40, z-score = -0.52, R) >droYak2.chr2L 1200491 120 - 22324452 UCGUUAUUAUUGCGCAAUAACAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUGAAUAUAUUGCUAGUCAGUCAAAAAAAUGUGGGUAUUCCCAAGCGUGAAUUGUGAAUCCGAUAAAUAA (((((((..(..(((......((((((((........))))))))....(((((.((......)).)))))..........((((....)))).)))..)...))))...)))....... ( -25.30, z-score = -1.56, R) >droSec1.super_14 1176772 111 - 2068291 UCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCCAAACUUAAUGUAUAGCUAGUAAGUUAAAAAAGUGUGGGUAUUCCCAGGCGUGAAUUGGGAAUAA--------- .........(..(((......((((((((........))))))))...((((((..(.....)...))))))......)))..).(((((((((.......))))))))).--------- ( -27.90, z-score = -2.29, R) >droSim1.chr2L 1197656 111 - 22036055 UCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAUGUAUAGCUAGUCAGUUAAAAAAGUGUGGGUAUUCCCAGGCGUGAAUUGGGAAUAA--------- .........(..(((......((((((((........))))))))..............(((((....))))).....)))..).(((((((((.......))))))))).--------- ( -26.40, z-score = -1.96, R) >consensus UCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAUGUAUUGCUAGUCAGUCAAAAAAGUGUGGGUAUUCCCAGGCGUGAAUUGGGAAUAA_________ .........(..(((......((((((((........))))))))....................................((((....)))).)))..).................... (-20.04 = -20.24 + 0.20)


| Location | 1,218,552 – 1,218,667 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Shannon entropy | 0.25194 |
| G+C content | 0.54949 |
| Mean single sequence MFE | -41.56 |
| Consensus MFE | -27.02 |
| Energy contribution | -29.10 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1218552 115 + 23011544 CGCCAGAUGGACCGAGCUGCCUUAGCUCGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUGGGCAAAU-----CUACUGGCCA .((((((((((((((((((...)))))))))......(((((.....)))))...(((((....)))))..)))).....(((..(((......))).)))....-----...))))).. ( -47.10, z-score = -3.59, R) >droEre2.scaffold_4929 1264130 107 + 26641161 ------------CAAUCUGCGUUAGCUUGGCAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUGG-CAAAUACUUGCUGCUCGCCA ------------......(((.(((.(((((.((.(((((((.....))))).(((((((....))))).))...)))).))))).))).....((..(-(((....))))..))))).. ( -36.70, z-score = -2.00, R) >droYak2.chr2L 1200611 110 + 22324452 ---------UGCCAGGUGGCGUUAGCUUCGUAGUAUCGCCCUUUCCAAGGGCAGCGAGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUGG-CAAAUAGUAGCUACUCUCCA ---------.(((....))).........(((((((((((((.....))))).(((.(((((............)))))))).))..)))))).(((((-(........))))))..... ( -32.30, z-score = -1.55, R) >droSec1.super_14 1176883 114 + 2068291 CGCCAGGUGGGCCGAGCUGCGUUAGCUCGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUGG-CAAAU-----CUACUGGCCA .(((.((((((((((((((...)))))))))....(((((((.....))))).(((((((....))))).))...)).)))))...........(((((-.....-----)))))))).. ( -46.90, z-score = -2.45, R) >droSim1.chr2L 1197767 114 + 22036055 CGCCAGGUGGGCCGAGCUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUGG-CAAAU-----CUACUGGCCA .(((.((((((((((((((...)))))))))....(((((((.....))))).(((((((....))))).))...)).)))))...........(((((-.....-----)))))))).. ( -44.80, z-score = -1.81, R) >consensus CGCCAG_UGGGCCGAGCUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUGG_CAAAU_____CUACUGGCCA .(((((....(((((((((...)))))))))(((.(((((((.....))))).(((.(((((............)))))))).))))).........................))))).. (-27.02 = -29.10 + 2.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:56 2011