
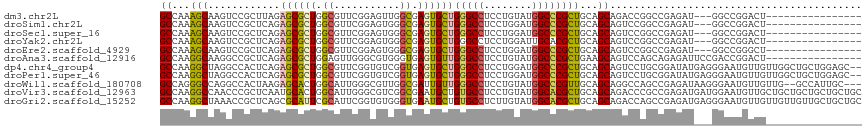
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,027,435 – 13,027,533 |
| Length | 98 |
| Max. P | 0.571995 |

| Location | 13,027,435 – 13,027,533 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.50748 |
| G+C content | 0.65039 |
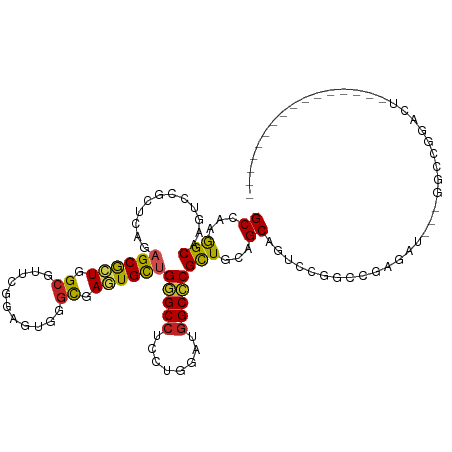
| Mean single sequence MFE | -49.58 |
| Consensus MFE | -24.77 |
| Energy contribution | -24.11 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 13027435 98 + 23011544 GCCAAAGCAAGUCCGCUUAGAGCGCUGGCGUUCGGAGUUGGCGAGUGCUGGGCCUCCUGUAUGGCCCGCUGCAGCAGACCGGCCGAGAU---GGCCGGACU---------------- .....((((.(((.((((.(((((....))))).)))).)))...))))(((((........)))))((....))...((((((.....---))))))...---------------- ( -45.00, z-score = -1.03, R) >droSim1.chr2L 12817802 98 + 22036055 GCCAAAGCAAGUCCGCUCAGAGCGCUGGCGUUCGGAGUGGGCGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCGGCCGAGAU---GGCCGGACU---------------- .....((((.((((((((.(((((....))))).))))))))...))))(((((........)))))((....))(((((((((.....---)))))))))---------------- ( -57.50, z-score = -3.42, R) >droSec1.super_16 1183427 98 + 1878335 GCCAAAGCAAGUCCGCUCAGAGCGCUGGCGUUCGGAGUGGGCGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCGGCCGAGAU---GGCCGGACU---------------- .....((((.((((((((.(((((....))))).))))))))...))))(((((........)))))((....))(((((((((.....---)))))))))---------------- ( -57.50, z-score = -3.42, R) >droYak2.chr2L 9446426 98 + 22324452 GCCAAAGCAAGUCCGCUCAGAGCGCUGGCGUUCGGAGUGGGCGAGUGCUGGGCCUCCUGGAUUGCACGCUGCAGCAGUCCGGCCGAGAU---GGCCGGACU---------------- ((...(((..((((((((.(((((....))))).))))))))..((((....((....))...)))))))...))(((((((((.....---)))))))))---------------- ( -54.40, z-score = -3.11, R) >droEre2.scaffold_4929 14224198 98 - 26641161 GCCAAAGCAAGUCCGCUCAGAGCGCUGGCGUUCGGAGUGGGCGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCGGCCGAGAU---GGCCGGGCU---------------- .....((((.((((((((.(((((....))))).))))))))...))))(((((........)))))((....))(((((((((.....---)))))))))---------------- ( -56.90, z-score = -2.86, R) >droAna3.scaffold_12916 3766557 101 - 16180835 GCCAAGGCAAGGCCGCUCAGAGCGCUGGAGUUGGGCGUGGGUGAGUGUUGGGCCUCCUGUAUGGCCCGCUGAAGCAGUCCAGCAGAGAUUCCGACCGGACU---------------- ((...(((.....((((((..(((((.......)))))...))))))..(((((........))))))))...))(((((.(..(......)..).)))))---------------- ( -37.30, z-score = 0.96, R) >dp4.chr4_group4 705416 115 + 6586962 GCCAAGGCUAGGCCACUCAGAGCGCUGGCGUUCGGUGUCGGUGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCUGCGGAUAUGAGGGAAUGUUGUUGGCUGCUGGAGC-- ((....))..(((.(((((..((((((.....))))).)..))))))))(((((........)))))(((.((((((((..(((.((((......))))))).)))))))).)))-- ( -50.10, z-score = -0.24, R) >droPer1.super_46 237477 115 - 590583 GCCAAGGCUAGGCCACUCAGAGCGCUGGCGUUCGGUGUCGGUGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCUGCGGAUAUGAGGGAAUGUUGUUGGCUGCUGGAGC-- ((....))..(((.(((((..((((((.....))))).)..))))))))(((((........)))))(((.((((((((..(((.((((......))))))).)))))))).)))-- ( -50.10, z-score = -0.24, R) >droWil1.scaffold_180708 6070708 112 + 12563649 GCCAGGGCCAGGCCACUAAGAGCACUGGCAUUGGGCGUUGGCGAUUGUUGGGCCUCCUGUAUGGCCCGUUGCAGCAGGCCAGCCGAGAUAAGGGAAUGUUGUUG--GCCAUUGC--- (((((.(((..((((..........))))....))).)))))(.((((((((((........))))))..))))).(((((((.((.((......)).))))))--))).....--- ( -46.30, z-score = -0.35, R) >droVir3.scaffold_12963 6883968 117 + 20206255 GCCAAGGCCAACCCGCUCAAUGCACUGGCAUUGGGCGUCGGCGAAUGCUGUGCCUCCUGUAUGGCACGCUGCAGCAGACCCGCCGAGAUGAUGGAAUGUUGCUGCUGCUGCUGCUGC (((..((....))((((((((((....))))))))))..)))....((.(((((........)))))((.(((((((.....(((......))).......))))))).)).))... ( -48.90, z-score = -0.96, R) >droGri2.scaffold_15252 10054546 117 + 17193109 GCCAAGGCUAAACCGCUCAGCGCAUUCGCAUUCGGUGUGGGUGAAUGCUGUGCCUCUUGUAUGGCACGCUGCAGCAGACCAGCCGAGAUGAGGGAAUGUUGUUGUUGUUGCUGCUGC ((((.((.....))((.((((.(((((((((...)))))))))...)))).))........))))..((.(((((((..((((.((.((......)).))))))...))))))).)) ( -41.40, z-score = 0.42, R) >consensus GCCAAGGCAAGUCCGCUCAGAGCGCUGGCGUUCGGAGUGGGCGAGUGCUGGGCCUCCUGGAUGGCCCGCUGCAGCAGUCCGGCCGAGAU___GGCCGGACU________________ ((...(((............((((((.((...........)).))))))(((((........))))))))...)).......................................... (-24.77 = -24.11 + -0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:42 2011