| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,027,288 – 13,027,382 |
| Length | 94 |
| Max. P | 0.978127 |

| Location | 13,027,288 – 13,027,382 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.34 |
| Shannon entropy | 0.26936 |
| G+C content | 0.53865 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -25.43 |
| Energy contribution | -24.99 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
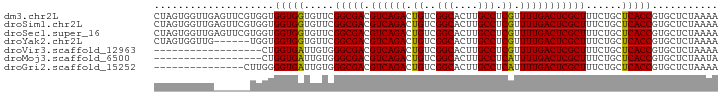
>dm3.chr2L 13027288 94 + 23011544 CUAGUGGUUGAGUUCGUGGUGGUGGUGUUCGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA .........((((...((((((..(.....(((((.((((((.((..(((....))).)).)))))))))))..)..).))))).))))..... ( -32.10, z-score = -1.50, R) >droSim1.chr2L 12817655 94 + 22036055 CUAGUGGUUGAGUUCGUGGUGGUGGUGUUCGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA .........((((...((((((..(.....(((((.((((((.((..(((....))).)).)))))))))))..)..).))))).))))..... ( -32.10, z-score = -1.50, R) >droSec1.super_16 1183280 94 + 1878335 CUAGUGGUUGAGUUCGUGGUGGUGGUGUUCGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA .........((((...((((((..(.....(((((.((((((.((..(((....))).)).)))))))))))..)..).))))).))))..... ( -32.10, z-score = -1.50, R) >droYak2.chr2L 9446285 88 + 22324452 CUAGUGGUUG------UGGUGGUGGUGUUCGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA .(((.(((..------.(((((..(.....(((((.((((((.((..(((....))).)).)))))))))))..)..).))))..))))))... ( -29.70, z-score = -1.59, R) >droVir3.scaffold_12963 6883821 76 + 20206255 ------------------CUGGUGAUUGUGGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA ------------------..(((((..(..(((((.((((((.((..(((....))).)).)))))))))))..)...)))))........... ( -27.40, z-score = -2.98, R) >droMoj3.scaffold_6500 26062290 76 + 32352404 ------------------CUGGUGAUUGUGGGCGACGUCAGACUGUCGGCACUUGCCUCAUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAUA ------------------..(((((..(..(((((.((((((.((..(((....))).)).)))))))))))..)...)))))........... ( -28.10, z-score = -3.66, R) >droGri2.scaffold_15252 10054387 79 + 17193109 ---------------CUUGGGGUGAUUGUGGGCGACGUCAGACUGUCGGCACUUGCCUCAUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA ---------------.....(((((..(..(((((.((((((.((..(((....))).)).)))))))))))..)...)))))........... ( -28.10, z-score = -2.84, R) >consensus CUAGUGGUUG______UGGUGGUGGUGUUCGGCGACGUCAGACUGUCGGCACUUGCCUCGUUUUGACUCGCUUUCUGCUCACCGUGCUCUAAAA ....................(((((.....(((((.((((((.((..(((....))).)).)))))))))))......)))))........... (-25.43 = -24.99 + -0.45)

| Location | 13,027,288 – 13,027,382 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Shannon entropy | 0.26936 |
| G+C content | 0.53865 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -19.51 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.853991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
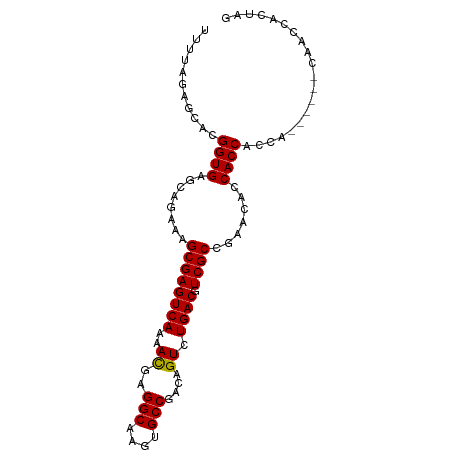
>dm3.chr2L 13027288 94 - 23011544 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCGAACACCACCACCACGAACUCAACCACUAG .....(((...((((........((((((((..((..(((....)))....)).)))).))))..........)))).....)))......... ( -20.27, z-score = -0.10, R) >droSim1.chr2L 12817655 94 - 22036055 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCGAACACCACCACCACGAACUCAACCACUAG .....(((...((((........((((((((..((..(((....)))....)).)))).))))..........)))).....)))......... ( -20.27, z-score = -0.10, R) >droSec1.super_16 1183280 94 - 1878335 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCGAACACCACCACCACGAACUCAACCACUAG .....(((...((((........((((((((..((..(((....)))....)).)))).))))..........)))).....)))......... ( -20.27, z-score = -0.10, R) >droYak2.chr2L 9446285 88 - 22324452 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCGAACACCACCACCA------CAACCACUAG ...........((((.(......((((((((..((..(((....)))....)).)))).))))......)))))....------.......... ( -20.10, z-score = -0.74, R) >droVir3.scaffold_12963 6883821 76 - 20206255 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCCACAAUCACCAG------------------ ...........(((((.......((((((((..((..(((....)))....)).)))).))))......)))))..------------------ ( -22.82, z-score = -2.31, R) >droMoj3.scaffold_6500 26062290 76 - 32352404 UAUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAAUGAGGCAAGUGCCGACAGUCUGACGUCGCCCACAAUCACCAG------------------ ...........(((((.......((((((((...((.(((....)))..))...)))).))))......)))))..------------------ ( -23.82, z-score = -3.10, R) >droGri2.scaffold_15252 10054387 79 - 17193109 UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAAUGAGGCAAGUGCCGACAGUCUGACGUCGCCCACAAUCACCCCAAG--------------- ...........(((((.......((((((((...((.(((....)))..))...)))).))))......))))).....--------------- ( -22.92, z-score = -2.58, R) >consensus UUUUAGAGCACGGUGAGCAGAAAGCGAGUCAAAACGAGGCAAGUGCCGACAGUCUGACGUCGCCGAACACCACCACCA______CAACCACUAG ...........((((........((((((((..((..(((....)))....)).)))).)))).......)))).................... (-19.51 = -19.30 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:41 2011