| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,016,086 – 13,016,205 |
| Length | 119 |
| Max. P | 0.652047 |

| Location | 13,016,086 – 13,016,205 |
|---|---|
| Length | 119 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.16 |
| Shannon entropy | 0.80826 |
| G+C content | 0.46913 |
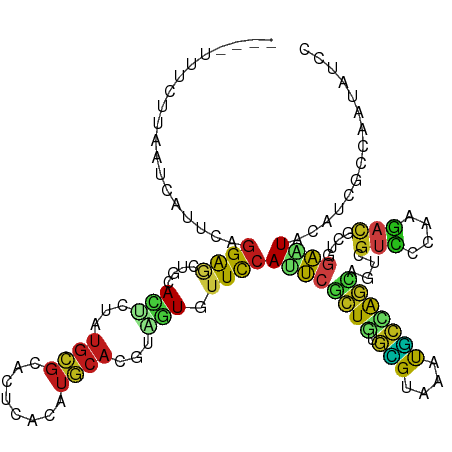
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -10.52 |
| Energy contribution | -9.70 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
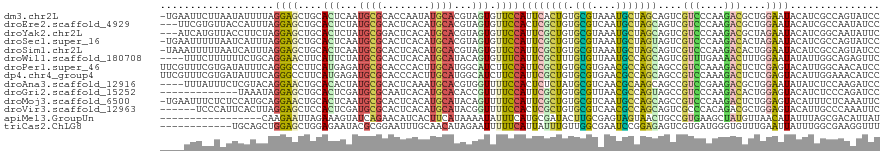
>dm3.chr2L 13016086 119 - 23011544 -UGAAUUCUUAAUAUUUUAGGAGCUGCACUCAAUGCGCACCAAUAUGCACGUAGUGUUCCAUUCACUGUGCGUAAAUGCUAGCAGUCGUCCCAAGACGCUGGAAUACAUCGCCAGUAUCC -.((((((((((....))))))(..(((((..(((.(((......))).))))))))..)))))((((.(((......(((((.(((.......)))))))).......))))))).... ( -32.92, z-score = -1.96, R) >droEre2.scaffold_4929 14212770 117 + 26641161 ---UUCGUGUUACCAUUUAGGAGCUGCACUCUAUGCGCACUCACAUGCACGUAGUGUUCCACUCGCUGUGCGUCAAUGCUAGCAGUCGUCCCAAGACGCUGGAAUACAUCGCCAAUAUCC ---..(((((..........(((.(((.(.....).))))))....)))))..(((((((....((((.(((....)))))))((.((((....)))))))))))))............. ( -31.44, z-score = -0.53, R) >droYak2.chr2L 9435066 117 - 22324452 ---AUCAUGUUACCUUCUAGGAGCUGCACUCUAUGCGGACUCACAUGCACGUAGUGUUCCAUUCGCUGUGCGUAAAUGCUAGCAGUCGUCCCAAGACGCUAGAAUACAUCGGCAAUAUUC ---...(((((.((......((((((((.....))))).)))..((((((..((((.......)))))))))).....(((((.(((.......))))))))........)).))))).. ( -31.30, z-score = -0.74, R) >droSec1.super_16 1171623 119 - 1878335 -UGAAUUUUUAAUCAUUUAGGAGCUGCACUCAAUGCGCACUCACAUGCACGUAGUGUUCCAUUCGCUGUGCGUAAAUGCUAGUAGUCGUCCCAAGACACUAGAAUACAUCGCCAGUAUCC -..................(((...(((((..(((.(((......))).)))))))))))....((((.(((......(((((.(((.......)))))))).......))))))).... ( -29.82, z-score = -1.51, R) >droSim1.chr2L 12809066 119 - 22036055 -UAAAUUUUUAAUCAUUUAGGAGCUGCACUCAAUGCGCACUCACAUGCACGUAGUGUUCCAUUCGCUGUGCGUAAAUGCUAGCAGUCGUCCCAAGACACUGGAAUACAUCGCCAGUAUCC -..................((((((((.....(((.(((......))).))).((((((((...((((.(((....))))))).(((.......)))..))))))))...).)))).))) ( -30.20, z-score = -1.41, R) >droWil1.scaffold_180708 6058753 116 - 12563649 ----UUUCUUUUUUCUGCAGGAACUUCAUUCUAUGCGCACUCACAUGCAUACAGUGUUUCAUUCGCUUUGUGUUAAUGCCAGCAGUCGUUUGAAAACUUUGGAAUAUAUUGGCAGAGUUC ----........((((((........((((.((((((........)))))).))))(((((..(((((((.((....))))).)).))..)))))................))))))... ( -23.20, z-score = 0.35, R) >droPer1.super_46 220820 120 + 590583 UUCGUUUCGUGAUAUUUCAGGGCCUUCAUGAGAUGCGCACCCACUUGCAUGGCAUCUUCCAUUCGCUGUGCGUGAACGCCAGCAGCCGUCCAAAGACUCUCGAGUACAUUGGCAACAUCC ...((((((.((........(((......((((((((((......)))...)))))))......((((.(((....))))))).)))(((....))))).)))).))..((....))... ( -35.40, z-score = -1.05, R) >dp4.chr4_group4 688413 120 - 6586962 UUCGUUUCGUGAUAUUUCAGGGCCUUCAUGAGAUGCGCACCCACUUGCAUGGCAUCUUCCAUUCGCUGUGCGUGAACGCCAGCAGCCGUCCAAAGACUCUCGAGUACAUUGGAAACAUCC ...((((((((.(((((.(((((......((((((((((......)))...)))))))......((((.(((....))))))).)))(((....))).)).))))))))..))))).... ( -37.10, z-score = -1.65, R) >droAna3.scaffold_12916 3755245 116 + 16180835 ----UUUAUUUCUCGUACAGGAACUGCACACUAUGCGCACUCAAAUGCACGUGGUUUUCCACUCUCUAUGCGUCAACGCAAGCAGCCGUCCGAAGACGCUGGAAUAUAUCUCCAAGAUCC ----..((((((.(((...(((.((((..((((((.(((......))).)))))).............((((....)))).))))...)))....)))..)))))).............. ( -30.00, z-score = -1.93, R) >droGri2.scaffold_15252 10041410 107 - 17193109 -------------UAAAUAGGAGCUGCACUCGAUGCGCAAUCACAUGCACACCGUUUUCCAUUCGCUGUGCGUUAACGCCAGUAGCCGUCCCAAGACACUGGAGUACAUCUCCCAGAUCC -------------......((((.((.((.((.((.(((......))).)).))...((((...((((.(((....)))))))....(((....)))..)))))).)).))))....... ( -27.30, z-score = -1.27, R) >droMoj3.scaffold_6500 26049621 119 - 32352404 -UGAAUUUCUCUCCAUGCAGGAACUGCACUCAAUGCGCACUCACAUGCAUACAGUUUUCCAUUCGCUGUGCGUCAAUGCCAGCAGCCGUCCCAAGACUCUGGAGUACAUUUCUCAAAUUC -.(((.....(((((.((.((((..(((.....)))(((......)))........))))....((((.(((....))))))).)).(((....)))..))))).....)))........ ( -27.80, z-score = -0.88, R) >droVir3.scaffold_12963 6871363 114 - 20206255 ------UCCCAUUCACUUAGGAGCUCCACUCGAUGCGCACUCACAUGCAUACGGUUUUCCACUCGCUGUGCGUCAACGCCAGCAGUCGCCCACAGACGCUGGAGUACAUUGCCCAAAUUC ------.............((.((.......((((((((......)))..(((((.........)))))))))).((.(((((.(((.......)))))))).)).....))))...... ( -30.10, z-score = -1.20, R) >apiMel3.GroupUn 374079202 103 + 399230636 -----------------CAAGAAUUAGAAAGUAUCAGAACAUCACUUCAUAAAAUAUUUCAUGCGAUACUUGCGAGUAGUAACUGCCGUGAAGCUAUGUUAACAUAUUUAGCGACAUUAU -----------------...........((((((...(((((..((((((............((((...))))(.((((...)))))))))))..)))))...))))))........... ( -16.00, z-score = 0.10, R) >triCas2.ChLG8 7722768 108 - 15773733 ------------UGCAGCUGGAGCUGGAGAAUACGCGGAAUUUGCAACAUAGAAUUUUUCAUUAUUUGUUGGCGAAUCCGGAGAGUCGUGAUGGGUGUUUGAAUUAUUUGGCGAAGGUUU ------------....((..((......((((((.((((.(((((((((...(((.....)))...)))).)))))))))(.....).......))))))......))..))........ ( -22.60, z-score = 0.78, R) >consensus ____UUUCUUAAUCAUUCAGGAGCUGCACUCUAUGCGCACUCACAUGCACGUAGUGUUCCAUUCGCUGUGCGUAAAUGCCAGCAGUCGUCCCAAGACGCUGGAAUACAUCGCCAAUAUCC ...................((((....(((...((((........))))...))).))))((((((((.(((....)))))))....(((....)))....))))............... (-10.52 = -9.70 + -0.82)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:37 2011