| Sequence ID | dm3.chr2L |
|---|---|
| Location | 13,013,809 – 13,013,918 |
| Length | 109 |
| Max. P | 0.630357 |

| Location | 13,013,809 – 13,013,918 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.32 |
| Shannon entropy | 0.58819 |
| G+C content | 0.45661 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -13.94 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
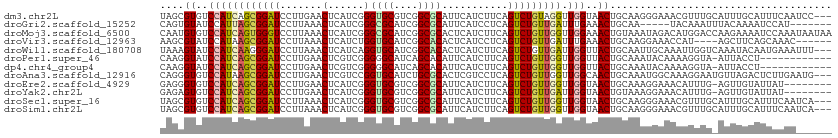
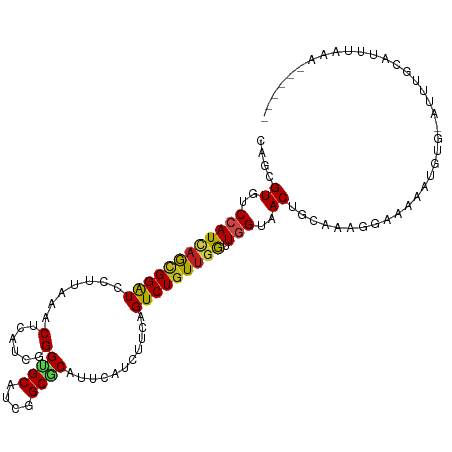
>dm3.chr2L 13013809 109 + 23011544 UAGCGUGUCCAUCAGCGGAUCCUUGAACUCAUCGGGUGCGUCGGCGCAUUCAUCUUCAGUCUGUAGGUUGGUAACUGCAAGGGAAACGUUUGCAUUUGCAUUUCAAUCC--- ..((((..(((((.((((((...((((......(((((((....)))))))...)))))))))).)).)))..))(((((((....).))))))...))..........--- ( -29.30, z-score = -0.39, R) >droGri2.scaffold_15252 10039349 100 + 17193109 CAGUGUAUCCAUUAGCGGAUCCUUAAACUCAUCGGGCGCAUCGGCGCAUUCAUCCUCAGUCUGUUGAUUUGAAACUGCAA-----UACAAAUUUACAAAAUCCAU------- ((((......((((((((((((...........))((((....))))...........)))))))))).....))))...-----....................------- ( -18.70, z-score = -0.45, R) >droMoj3.scaffold_6500 26047562 112 + 32352404 CAAUGUGUCCAUCAGUGGGUCCUUAAACUCAUCGGGCGCAUCGGCGCACUCAUCUUCAGUCUGUUGGUUGGAAACUGUAAAUAGACAUGGACCAAGAAAAUCCAAAUAAUAA ...((.(((((((.((((((......)))))).))((((....))))...........(((((((....(....)....))))))).))))))).................. ( -29.90, z-score = -1.79, R) >droVir3.scaffold_12963 6868822 102 + 20206255 AAGCGUAUCCAUAAGCGGAUCCUUAAACUCAUCUGGUGCAUCGGCACACUCAUCCUCAGUCUGUUGAUUUGAAACUGCAAGGAAACCAU----AGCUUCAGCAAAC------ (((.(.((((......))))))))............((((((((((.(((.......))).))))))).((((.(((...(....)..)----)).)))))))...------ ( -18.90, z-score = 0.47, R) >droWil1.scaffold_180708 6056466 109 + 12563649 UAAAGUAUCCAUCAAGGGAUCCUUAAACUCAUCAGGUGCAUCGGCACACUCAUCUUCAGUCUGUUGAUUGGUUACUGCAAUUGCAAAUUGGUCAAAUACAAUGAAAUUU--- ..(((.((((......)))).)))...........((((....)))).......((((.....(((((..(((..(((....))))))..)))))......))))....--- ( -23.80, z-score = -1.35, R) >droPer1.super_46 218694 99 - 590583 CAAGGUAUCCAUCAGCGGAUCCUUGAACUCGUCGGGGGCAUCAGCACAUUCAUCUUCAGUCUGUUGGUUGGUUACUGCAAAUACAAAAGGUA-AUUACCU------------ ...((((.((((((((((((((((((.....))))))((....)).............))))))))).))).))))...........((((.-...))))------------ ( -26.50, z-score = -1.93, R) >dp4.chr4_group4 686287 99 + 6586962 CAAGGUAUCCAUCAGCGGAUCCUUGAACUCGUCGGGGGCAUCAGCACAUUCAUCUUCAGUCUGUUGGUUGGUUACUGCAAAUACAAAAGGUA-AUUACCU------------ ...((((.((((((((((((((((((.....))))))((....)).............))))))))).))).))))...........((((.-...))))------------ ( -26.50, z-score = -1.93, R) >droAna3.scaffold_12916 3753212 109 - 16180835 CAGGGUGUCCAUAAGCGGAUCCUUGAACUCGUCCGGUGCAUCUGCGCACUCGUCCUCAGUCUGUUGGUUGGCAACUGCAAAUGGCAAAGGAAUGUUAGACUCUUGAAUG--- (((((.((((......))))))))).....(((((((((......)))).))((((..(((.....((.(....).))....)))..))))......))).........--- ( -29.50, z-score = 0.31, R) >droEre2.scaffold_4929 14210527 103 - 26641161 GAGGGUGUCCAUCAGCGGAUCCUUGAACUCAUCGGGUGCGUCGGCGCAUUCAUCUUCAGUCUGUUGGUUGGUAACUGCAAAGGAAACAUUUG-AGUUGUAUUAU-------- ........((((((((((((...((((......(((((((....)))))))...))))))))))))).)))(((((.(((((....).))))-)))))......-------- ( -29.30, z-score = -0.80, R) >droYak2.chr2L 9432721 103 + 22324452 GAGAGUGUCCAUCAGCGGAUCCUUGAACUCAUCGGGUGCGUCGGCGCAUUCAUCUUCAGUCUGUUGAUUGGUAACUGUAAAGGAAACAUUUG-AGUUGUAUUAU-------- ..(((..(((......)))..))).((((((..(((((((....)))))))....((((((....))))))..........(....)...))-)))).......-------- ( -28.10, z-score = -0.82, R) >droSec1.super_16 1169298 109 + 1878335 UAGCGUGUCCAUCAGCGGAUCCUUAAACUCAUCGGGUGCGUCGGCGCAUUCAUCUUCAGUCUGUUGGUUGGUAACUGCAAGGGAAACGUUUGCAUUUGCAUUUCAAUCA--- ..((((..(((((((((((..............(((((((....)))))))..(....))))))))).)))..))(((((((....).))))))...))..........--- ( -29.60, z-score = -0.95, R) >droSim1.chr2L 12806766 109 + 22036055 UAGCGUGUCCAUCAGCGGAUCCUUAAACUCAUCGGGUGCGUCGGCGCAUUCAUCUUCAGUCUGUUGGUUGGUAACUGCAAGGGAAACGUUUGCAUUUGCAUUUCAAUCA--- ..((((..(((((((((((..............(((((((....)))))))..(....))))))))).)))..))(((((((....).))))))...))..........--- ( -29.60, z-score = -0.95, R) >consensus CAGCGUGUCCAUCAGCGGAUCCUUAAACUCAUCGGGUGCAUCGGCGCAUUCAUCUUCAGUCUGUUGGUUGGUAACUGCAAAGGAAAAAUGUG_AUUUGCAUUUAAA______ ....((..((((((((((((.......(......)((((....))))...........))))))))).)))..))..................................... (-13.94 = -13.88 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:37 2011