| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,977,316 – 12,977,419 |
| Length | 103 |
| Max. P | 0.606334 |

| Location | 12,977,316 – 12,977,419 |
|---|---|
| Length | 103 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.32137 |
| G+C content | 0.59452 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.07 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12977316 103 + 23011544 -----------------GUCGCGUUAAGUGCUUGGGUAACCCCGAGCGCGAGGGAUCCGUGUCCAUUGUCGGAGCUGUGUCUCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACCUCCGC -----------------(((((.....(((((((((....)))))))))..((((((.(.(((.((..((((((.........)))).))..)))))....).)))))))))))...... ( -43.80, z-score = -3.14, R) >droSim1.chr2L 12769087 102 + 22036055 -----------------GUCGCGU-AAGUGCUUGGGUAACCCCGAGCGCGAAGGAUCCGUGUCCAUUGUCGGAGCUGUGUCUCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACCUCCGC -----------------((((((.-..(((((((((....)))))))))...((((....))))...(((((((....(((.((.......)).)))..)))))))..))))))...... ( -42.80, z-score = -3.61, R) >droSec1.super_16 1131834 103 + 1878335 -----------------GUCGCGUUAAGUGCUUGGGUAACCCCGAGCGCGAGGGAUCCGUGUCCAUUGUCGGAGCUGUGUCCCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACCUCCGC -----------------(((((.....(((((((((....)))))))))..((((((...(........)((((..((..((((....)).))..))..)))))))))))))))...... ( -43.90, z-score = -3.24, R) >droYak2.chr2L 9395674 103 + 22324452 -----------------GUCGCGUCAAGUGCUUGGGUAACCCUGAGCGCGAGGGAUCCGUGUCCAUUGUCGGAGCUGUGUCUCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACAUCCGC -----------------(((((.....(((((..((....))..)))))..((((((.(.(((.((..((((((.........)))).))..)))))....).)))))))))))...... ( -42.20, z-score = -2.39, R) >droEre2.scaffold_4929 14171807 103 - 26641161 -----------------GUCGCGUUAAGUGCUUGGGUAACCCCGAGCGCGAGGGAUCCGUGUCCAUUGUCGGAGCUGUGUCUCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACAUCCGC -----------------(((((.....(((((((((....)))))))))..((((((.(.(((.((..((((((.........)))).))..)))))....).)))))))))))...... ( -44.70, z-score = -3.33, R) >droAna3.scaffold_12916 3716768 103 - 16180835 -----------------GUCGCGUCAAGUGCUUGGGUAACCCCGAGCGCGAGGGUUCCGUGUCCAUUGUCGGUGCUGUGUCGCCUCCUGGUGGUGAUUUCUCCGAUCCCGUCACCUCCGC -----------------...(((....(((((((((....)))))))))((((....((.(......(((((.(....((((((.......))))))..).)))))).))...))))))) ( -36.60, z-score = -1.11, R) >droWil1.scaffold_180708 6007738 103 + 12563649 -----------------GUCGUGUCAAGUGUUUGGGUAACCCUGAACGUGAGGGCUCCGUGUCCAUUGUCGGAGCUGUAUCGCCACCUGGUGGUGAUUUCUCUGAUCCCGUCACUUCUGC -----------------............(((..((....))..)))((((((((.....))))...(((((((....((((((((...))))))))..)))))))....))))...... ( -34.60, z-score = -1.54, R) >dp4.chr4_group4 644412 103 + 6586962 -----------------GUCGCAUCAAGUGCUUGGGUAACCCCGAGCGCGAGGGUUCCGUCUCUAUUGUCGGUGCUGUGUCGCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACAUCCGC -----------------(((((.....(((((((((....)))))))))((((((.(((.(......).))).)))..((((((.......))))))..))).......)))))...... ( -42.20, z-score = -2.80, R) >droPer1.super_46 177881 103 - 590583 -----------------GUCGCAUCAAGUGCUUGGGUAACCCCGAGCGCGAGGGUUCCGUCUCCAUUGUCGGUGCUGUGUCGCCUCCUGGUGGUGACUUCUCCGAUCCCGUGACAUCCGC -----------------(((((.....(((((((((....)))))))))((((((.(((.(......).))).)))..((((((.......))))))..))).......)))))...... ( -42.20, z-score = -2.56, R) >droVir3.scaffold_12963 6825343 103 + 20206255 -----------------GUCGUGUCAAGUGCUUGGGUAAUCCCGAGCGUGAGGGCUCCGUGUCCAUUGUCGGCGCUGUCUCGCCACCUGGUGGUGAUUUCUCCGAUCCCGUUACUUCCGC -----------------...(((......(((((((....)))))))((((((((.....))))...(((((.(..((((((((....))))).)))..).)))))....))))...))) ( -35.60, z-score = -1.29, R) >droMoj3.scaffold_6500 26006750 103 + 32352404 -----------------GUCGUGUCAAGUGCUUGGGUAAUCCCGAACGUGAGGGCUCCGUGUCCAUUGUCGGCGCUGUCUCGCCACCUGGUGGUGAUUUCUCCGAUCCCGUUACCUCCGC -----------------((.(((...((((((((((....))))).(((((((((.....)))).))).)))))))....))).))..((.((((((............)))))).)).. ( -29.60, z-score = 0.45, R) >droGri2.scaffold_15252 10002775 103 + 17193109 -----------------GUCGCAUCAAGUGUUUGGGUAAUCCCGAGCGUGAGGGUUCCGUGUCCAUUGUGGGUGCUGUCUCACCACCCGGUGGUGAUUUCUCCGAUCCCGUUACGUCCGC -----------------...((.......(((((((....)))))))(((((((.((.(..(((.....)))..)....(((((((...))))))).......)).))).))))....)) ( -35.20, z-score = -1.14, R) >anoGam1.chr2R 29338918 103 - 62725911 -----------------GUCGCGUCAAGUGUCUCGGCAAUCCGGAGCGCGAAGGUUCCGUGUCGAUCGUCGGUGCCGUCUCGCCGCCCGGUGGUGAUUUCUCCGAUCCCGUCACUUCCGC -----------------...(((..(((((.(((((((...((((((......))))))))))))..(((((.(..((((((((....))))).)))..).)))))...).))))).))) ( -38.10, z-score = -0.67, R) >apiMel3.Group9 9324576 120 - 10282195 AACAUUAUAUAUUGUUUGCAGAGUGAAAUGUUUAGGUAAUCCCGAUCGAGAAGGUUCCGUCAGUAUUGUAGGUGCUGUAUCACCACCAGGUGGUGAUUUCUCUGAUCCUGUUACUAGUGC ((((((.(((.(((....))).))).))))))..((((((...((((((((((.......((((((.....))))))..(((((((...))))))))))))).))))..))))))..... ( -33.20, z-score = -1.84, R) >triCas2.ChLG4 10026634 103 + 13894384 -----------------GGCGUGUUAAAUGCUUGGGUAACCCUGAUCGUGAAGGUUCCGUUUCUAUUGUCGGGGCCGUAUCGCCCCCUGGUGGUGAUUUCUCUGAUCCCGUCACCUCAGC -----------------............(((.((((.((...(((((.((((((((((...(....).)))))))..(((((((....).)))))))))..)))))..)).)))).))) ( -31.40, z-score = -0.64, R) >consensus _________________GUCGCGUCAAGUGCUUGGGUAACCCCGAGCGCGAGGGUUCCGUGUCCAUUGUCGGAGCUGUGUCGCCUCCUGGUGGUGAUUUCUCCGAUCCCGUGACCUCCGC ....................((.....(((((((((....)))))))))..(((.((.............((((......))))....((.((.....)).)))).))).........)) (-22.71 = -22.07 + -0.63)


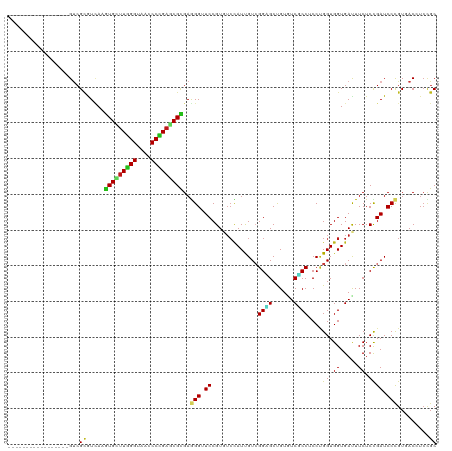
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:31 2011