| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,964,755 – 12,964,859 |
| Length | 104 |
| Max. P | 0.725404 |

| Location | 12,964,755 – 12,964,859 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.32594 |
| G+C content | 0.41259 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.38 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
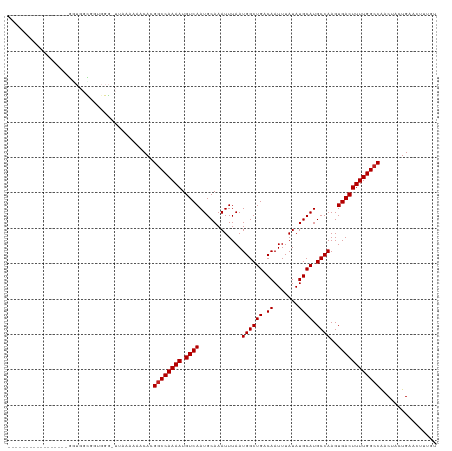
>dm3.chr2L 12964755 104 + 23011544 ----------------GGGUGGCGGUGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU ----------------((((..(((((((-((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..)))))).))))... ( -29.84, z-score = -1.68, R) >droSim1.chr2L 12756406 104 + 22036055 ----------------GGGUGGCGGUGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU ----------------((((..(((((((-((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..)))))).))))... ( -29.84, z-score = -1.68, R) >droSec1.super_16 1119484 104 + 1878335 ----------------GGGUGGCGGUGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU ----------------((((..(((((((-((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..)))))).))))... ( -29.84, z-score = -1.68, R) >droYak2.chr2L 9381890 120 + 22324452 AGGGUUGCGGGGUUGCGGGUGGCGGUGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU ......((((((((((.....))((((((-((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..))))).)))))))) ( -32.64, z-score = -0.03, R) >droEre2.scaffold_4929 14159405 106 - 26641161 --------------GCGGGUGGCGGUGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU --------------..((((..(((((((-((....)))..((((((((.((((............((((((.((.......)))).))))..))))))))))))..)))))).))))... ( -30.14, z-score = -1.33, R) >droAna3.scaffold_12916 3705687 118 - 16180835 ---AAAAGGAUGCCUGAGCUGGCGCCGGGGUUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU ---....((.((((......))))))((((((.........((((((((.((((............((((((.((.......)))).))))..))))))))))))........)))))).. ( -31.07, z-score = -0.83, R) >dp4.chr4_group4 620655 103 + 6586962 -----------------GAUGGUAGGGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCGACUACUGAACUUUGU -----------------...(((((...(-((....))).(((((((((.((((............((((((.((.......)))).))))..))))))))))))).)))))......... ( -27.94, z-score = -2.02, R) >droPer1.super_46 154173 103 - 590583 -----------------GAUGGUAGGGGG-UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU -----------------...(((((..((-((((((..((((((......))).............((((((.((.......)))).)))).)))..))))))))..)))))......... ( -26.60, z-score = -1.54, R) >droWil1.scaffold_180708 5992899 104 + 12563649 -----------------AGAGGUUAGGGAGGUGGAAAACACGACCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUGCUGAACUUUGU -----------------.((((((.((.(((((.....)))(.((((((.((((............((((((.((.......)))).))))..)))))))))).)..)).)).)))))).. ( -24.94, z-score = -0.62, R) >droVir3.scaffold_12963 13394300 90 - 20206255 -------------------------------GCUGCUGCACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUUC -------------------------------((....))..((((((((.((((............((((((.((.......)))).))))..))))))))))))................ ( -22.44, z-score = -1.60, R) >droMoj3.scaffold_6500 6359116 90 - 32352404 -------------------------------GAUGCUGCACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUUC -------------------------------..........((((((((.((((............((((((.((.......)))).))))..))))))))))))................ ( -20.94, z-score = -1.31, R) >droGri2.scaffold_15252 9991100 90 + 17193109 -------------------------------GACGCUGCACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUUC -------------------------------..........((((((((.((((............((((((.((.......)))).))))..))))))))))))................ ( -20.94, z-score = -1.51, R) >consensus _________________GGUGGCGGUGGG_UUAAAAAACACGGCCAAAAUGUCAAUGCAAAUUUAAUGGCUGACAAUUUAUAUGCAUGCCAAGUGACUUUUGGCCAACUACUGAACUUUGU .........................................((((((((.((((............((((((.((.......)))).))))..))))))))))))................ (-20.30 = -20.38 + 0.08)


| Location | 12,964,755 – 12,964,859 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.32594 |
| G+C content | 0.41259 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.13 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
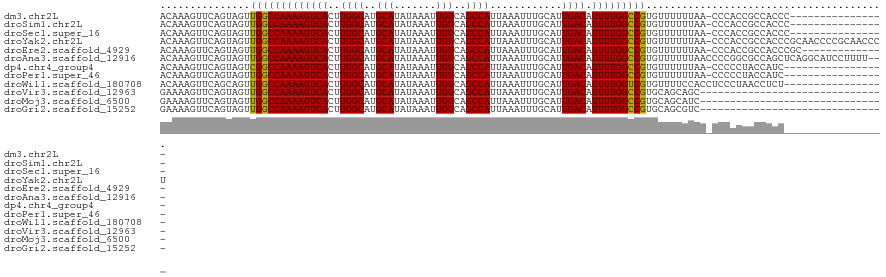
>dm3.chr2L 12964755 104 - 23011544 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCACCGCCACCC---------------- ..........(..((.((((((((((((..((((..(((.......)))..))))............)))).))))))))(.(((....))-).)))..).....---------------- ( -21.44, z-score = -0.40, R) >droSim1.chr2L 12756406 104 - 22036055 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCACCGCCACCC---------------- ..........(..((.((((((((((((..((((..(((.......)))..))))............)))).))))))))(.(((....))-).)))..).....---------------- ( -21.44, z-score = -0.40, R) >droSec1.super_16 1119484 104 - 1878335 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCACCGCCACCC---------------- ..........(..((.((((((((((((..((((..(((.......)))..))))............)))).))))))))(.(((....))-).)))..).....---------------- ( -21.44, z-score = -0.40, R) >droYak2.chr2L 9381890 120 - 22324452 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCACCGCCACCCGCAACCCCGCAACCCU .............(((((((((.......))))).(((....(((.((((((.((.......))..)))))).)))(((.(((........-..))).)))....)))......))))... ( -24.10, z-score = -0.31, R) >droEre2.scaffold_4929 14159405 106 + 26641161 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCACCGCCACCCGC-------------- ..........(..((.((((((((((((..((((..(((.......)))..))))............)))).))))))))(.(((....))-).)))..).......-------------- ( -21.44, z-score = 0.02, R) >droAna3.scaffold_12916 3705687 118 + 16180835 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAACCCCGGCGCCAGCUCAGGCAUCCUUUU--- .....(((..(.((((((((((((((((..((((..(((.......)))..))))............)))).)))))((((.(((....)))..)))))))))))).)))........--- ( -29.14, z-score = -0.83, R) >dp4.chr4_group4 620655 103 - 6586962 ACAAAGUUCAGUAGUCGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCCCUACCAUC----------------- ..........((((.(((((((((((((..((((..(((.......)))..))))............)))).))))))))).(((....))-)...))))....----------------- ( -24.64, z-score = -2.36, R) >droPer1.super_46 154173 103 + 590583 ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA-CCCCCUACCAUC----------------- ..........((((..((((((((((((..((((..(((.......)))..))))............)))).))))))))..(((....))-)...))))....----------------- ( -23.24, z-score = -1.57, R) >droWil1.scaffold_180708 5992899 104 - 12563649 ACAAAGUUCAGCAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGUCGUGUUUUCCACCUCCCUAACCUCU----------------- .........((((..(((((((((((((..((((..(((.......)))..))))............)))).)))))))))))))...................----------------- ( -19.54, z-score = -0.29, R) >droVir3.scaffold_12963 13394300 90 + 20206255 GAAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGCAGCAGC------------------------------- ..........((.(((((((((((((((..((((..(((.......)))..))))............)))).))))))))).)).))...------------------------------- ( -23.44, z-score = -0.97, R) >droMoj3.scaffold_6500 6359116 90 + 32352404 GAAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGCAGCAUC------------------------------- ..........((.(((((((((((((((..((((..(((.......)))..))))............)))).))))))))).)).))...------------------------------- ( -23.44, z-score = -1.36, R) >droGri2.scaffold_15252 9991100 90 - 17193109 GAAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGCAGCGUC------------------------------- ..........((.(((((((((((((((..((((..(((.......)))..))))............)))).))))))))).)).))...------------------------------- ( -23.44, z-score = -1.27, R) >consensus ACAAAGUUCAGUAGUUGGCCAAAAGUCACUUGGCAUGCAUAUAAAUUGUCAGCCAUUAAAUUUGCAUUGACAUUUUGGCCGUGUUUUUUAA_CCCACCGCCACC_________________ ...............(((((((((((((..((((..(((.......)))..))))............)))).)))))))))........................................ (-21.28 = -21.13 + -0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:30 2011