| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,926,903 – 12,927,015 |
| Length | 112 |
| Max. P | 0.516411 |

| Location | 12,926,903 – 12,927,015 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 69.22 |
| Shannon entropy | 0.67590 |
| G+C content | 0.50034 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -14.83 |
| Energy contribution | -13.23 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.88 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.516411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
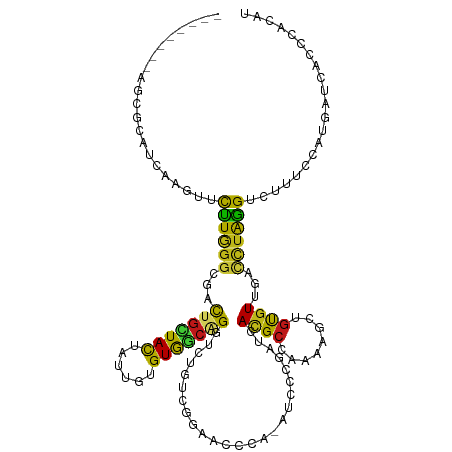
>dm3.chr2L 12926903 112 + 23011544 --------AGCGCAUCAAGUUCCUGGGGGACUGCUACUACUGUGUGGCGGGUCUGUCGGACCCCG-AUCCCGAUCACGCCAAUAACUGUGUUAUCCUAGGUCUUUCCAUGAUCAACCACAU --------....(((.(((..(((((((((..(((((......)))))(((((.....)))))..-.))))(((.((((........)))).)))))))).)))...)))........... ( -33.80, z-score = -0.35, R) >droSim1.chr2L 12722046 112 + 22036055 --------AGCGCAUCAAGUUCCUGGGGGACUGCUACUAUUGUGUGGCGGGUCUGUCGGACCCCG-AUCCCGAUCACGCCAAUAACUGUGUUAUCCUAGGUCUUUCCAUGAUUAACCACAU --------....(((.(((..(((((((((..(((((......)))))(((((.....)))))..-.))))(((.((((........)))).)))))))).)))...)))........... ( -33.80, z-score = -0.56, R) >droSec1.super_16 1088838 112 + 1878335 --------AGCGCAUCAAGUUCCUGGGGGACUGCUACUACUGUGUGGCGGGUCUGUCGGACCCCG-AUCCCGAUCACGCCAAUAACUGUGUUAUCCUAGGUCUUUCCAUGAUCAACCACAU --------....(((.(((..(((((((((..(((((......)))))(((((.....)))))..-.))))(((.((((........)))).)))))))).)))...)))........... ( -33.80, z-score = -0.35, R) >droYak2.chr2L 9349568 112 + 22324452 --------AGCGCAUCAAGUUUCUGGGCGACUGCUACUAUUGUGUGGCCGGUCUGUCGGAACCAG-AUCCCGAUCACGCCGACAACUGUGUUUCUCUAGGCCUUUUCAUGAUCACCCACAU --------...(.((((.(.....(((((((((((((......)))).)))))((((((.....(-((....)))...))))))...............))))...).)))))........ ( -29.20, z-score = 0.28, R) >droEre2.scaffold_4929 14128064 112 - 26641161 --------AGCGCAUCAAGUUCCUGGGGGACUGCUACUAUUGUGUGGCGGGUCUGUCCGAACCCG-AUCCCGAUCACGCCACCAACUGUGUUUCCCUAGGCCUUUCCAUGAUCACCAACAU --------....(((.(((..(((((((((..((.((..(((.(((((((((........))))(-((....)))..))))))))..))))))))))))).)))...)))........... ( -34.80, z-score = -0.88, R) >droAna3.scaffold_12916 3675882 112 - 16180835 --------AGCGGAUCAAGUUUCUUGGGGAUUGUUACUAUUGCGUGGCUGGUUUGGUUGAACCGG-AUCCGGAUCAUGCUGACAACUGUGUGGGCCUGGGUCUGUGCAUGAUCUCCGAGAU --------.............(((((((((((((((.....(((((((((((((((.....))))-).)))).))))))))))...((((..(((....)))..)))).))))))))))). ( -42.10, z-score = -2.35, R) >dp4.chr4_group4 577275 112 + 6586962 --------AGCGAAUCAAGUUCUUGGGCGACUGUUACUACUGUGUGGCAGGUCUAACGCCGCCGA-GUCCCGACCACGCCAAGAGUUGUGUAGGUCUGGGUCUGUCCAUGAUCUCCCAUAU --------.........((.((.((((((((((((((......)))))).)))...........(-(.(((((((((((........)))).)))).))).))))))).)).))....... ( -34.20, z-score = -0.07, R) >droPer1.super_46 112447 112 - 590583 --------AGCGAAUCAAGUUCUUGGGCGACUGCUACUACUGUGUGGCAGGUCUAACGCCGCCGA-GUCCCGACCACGCCAAGAGUUGUGUAGGUCUGAGUCUGUCCAUGAUCACCCACAU --------..........(.((.((((((((((((((......)))))).)))..........((-.((..((((((((........)))).)))).)).)).))))).)).)........ ( -32.50, z-score = 0.37, R) >droWil1.scaffold_180708 11725673 112 + 12563649 --------AACGUAUUAAGUUCUUGGGCGAUUGCUACUAUUGUGUAGCAGGUCUAAUACGUCCCA-AUCCCGACCAUGCAAAAUGCUGUGUUGACCUAGGCCUCUCAAUGAUCGCUCACAU --------...............((((((((((((((......)))))(((((.((((((((...-.....)))...((.....)).))))))))))............)))))))))... ( -32.80, z-score = -2.61, R) >droVir3.scaffold_13049 8008794 112 - 25233164 --------AACGUAUCAAAUUCUUAGGUGACUGCUACUAUUGUGUGGCAGGACUAUCAAGUCCCA-ACCCGGAUCACGCCAAAUGUUGCAUAGACCUGGGCCACUGUAUGAUCGCGCAUAU --------..(((((((......(((....)))..........(((((.(((((....)))))..-..((((.((..((........))...)).)))))))))....)))).)))..... ( -28.90, z-score = -0.20, R) >droGri2.scaffold_15110 14963171 112 + 24565398 --------AACGCAUCAAAUUCUUAGGUGACUGCUACUAUUGUGUGGCAGGUCUGACACGACCUA-AUCCGGAUCAUGCCAAGUGUUGUGUCGAACUGGGACAUUGUAUGAUUGCAAACAU --------...((((((.((((((((..(((((((((......)))))).)))(((((((((((.-....((......)).)).)))))))))..))))))....)).))).)))...... ( -32.90, z-score = -1.31, R) >droMoj3.scaffold_6680 23047979 112 + 24764193 --------AGCGAAUCAAAUUCUUAGGAGACUGCUACUAUUGUGUAGCGGGUCUUAUAAACCCAG-AUCCUAACCAUGCCAAAUGCUGCGUGGAUUUAGGUCUUUGUAUGAUCGAUAGUAU --------..(((.(((......(((((..(((((((......))))))).)))))...((..((-(.(((((((((((........))))))..))))))))..)).))))))....... ( -32.70, z-score = -2.34, R) >apiMel3.Group7 9129202 101 - 9974240 --------UGCGUAUAAAAUUUUUGGGCGACUGUUAUUAUUGUGUAAGCGGUGUGCCGACACCAA-AUUCUCAACAUGCGAAAAGCUGCGUGGAUUUAGGUAG--AUACGAU--------- --------..(((((..........((((((((((((......)).)))))).))))...(((((-(((.....((((((......))))))))))).)))..--)))))..--------- ( -23.50, z-score = 0.11, R) >triCas2.ChLG5 13822473 120 - 18847211 UGACUGUUUGCGAAUCAAACUUUUGGGUGACUGUUACUACUGCGUGUCAGGUUUGCCGA-ACGCACGCGAGGACCAUGCAGACUGUGCCGUCGAAAUGGGCCUGAACAUGAUCAAAGCAAU .......((((..((((.......((....))(((.....(((((((..((....))..-..)))))))(((.((((...(((......)))...)))).))).))).))))....)))). ( -32.70, z-score = 0.31, R) >anoGam1.chr2L 47917282 112 + 48795086 --------UGCGGAUAAAGUUCCUGGGCGAUUGCUACUACUGCGUGUCGGGCGUACCGGUGCGCA-AUAAGUAUCACGCCAAAAGCUGCGUCAAUUUGGGGUUGCGCAUGAUCAAGGAUAU --------.(((((......)))..((((..((.((((..((((..((((.....))))..))))-...)))).))))))....))((((.((((.....))))))))............. ( -37.70, z-score = -0.67, R) >consensus ________AGCGCAUCAAGUUCUUGGGCGACUGCUACUAUUGUGUGGCAGGUCUGUCGGAACCCA_AUCCCGAUCACGCCAAAAGCUGUGUUGACCUAGGUCUUUCCAUGAUCACCCACAU .....................((((((...(((((((......))))))).........................((((........))))...))))))..................... (-14.83 = -13.23 + -1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:25 2011