| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,907,641 – 12,907,732 |
| Length | 91 |
| Max. P | 0.936882 |

| Location | 12,907,641 – 12,907,732 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.55201 |
| G+C content | 0.55768 |
| Mean single sequence MFE | -17.18 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
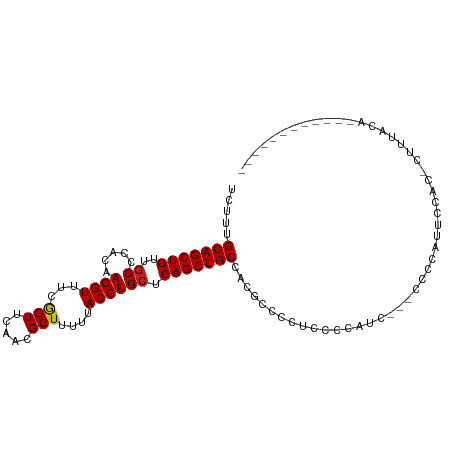
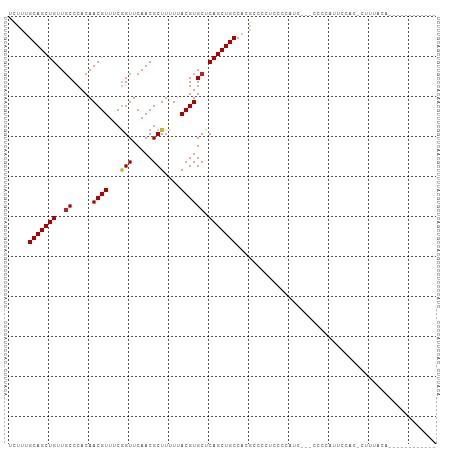
>dm3.chr2L 12907641 91 + 23011544 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCAGCUCCACAUC---CCCCAUUCCAC-CUUUACA------------ .....(((((((..((.....((((...(((.....)))....)))))).)))))))................---...........-.......------------ ( -16.20, z-score = -1.20, R) >droSim1.chr2L 12702257 90 + 22036055 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCAGCUCCACAUC----CCCAUUCCAC-CUUUACA------------ .....(((((((..((.....((((...(((.....)))....)))))).)))))))................----..........-.......------------ ( -16.20, z-score = -1.13, R) >droSec1.super_16 1067994 90 + 1878335 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCAGCUCCACAUC----CCCAUUCCAC-CUUUACA------------ .....(((((((..((.....((((...(((.....)))....)))))).)))))))................----..........-.......------------ ( -16.20, z-score = -1.13, R) >droYak2.chr2L 9328866 94 + 22324452 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCAGCUCCACAUCACAUCCCCUUCCAC-CUUUAAA------------ .....(((((((..((.....((((...(((.....)))....)))))).)))))))..............................-.......------------ ( -16.20, z-score = -1.06, R) >droEre2.scaffold_4929 14105308 91 - 26641161 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCAGCUCCACCUC---CCCCUAUCCAC-CUUUGCA------------ ....((((((.((((.((..........))..)))))).......(((...((((((...)))))).)))...---...........-...))))------------ ( -17.30, z-score = -0.99, R) >droAna3.scaffold_12916 3656496 82 - 16180835 UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCUCCACCUCC----AGCCUUGAA--------------------- .....(((((((..((.....((((...(((.....)))....)))))).)))))))................----.........--------------------- ( -16.20, z-score = -0.35, R) >dp4.chr4_group4 564338 95 + 6586962 UUUCCGCAGCUGUUUCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCCUCGCCAUC--CCCCCC----------GGCAGCAACUUCUAUA .....(((((((.........((((...(((.....)))....))))...)))))))..........(((...--......----------)))............. ( -18.00, z-score = -1.06, R) >droPer1.super_46 99539 105 - 590583 UUUCCGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCCUCGCCAUC--CCCCCCCCCAACACCCAACAGCAACUUCUAUA .....(((((((..((.....((((...(((.....)))....)))))).)))))))................--................................ ( -16.50, z-score = -1.36, R) >droWil1.scaffold_180708 5950178 93 + 12563649 UAUUUGCAGCUGUUGCCCACAACGUUCCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACACCCCCCGCAAUU--GCCGCACCCCACCUCCGGCA------------ .....(((((((..((.....((((...(((.....)))....)))))).)))))))...............(--((((...........)))))------------ ( -21.20, z-score = -0.90, R) >droMoj3.scaffold_6500 25949629 79 + 32352404 UCCCUGCAGCUGUUGCCCACAACGUUCCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCACCUCACCCUAGCCA---------------------------- .....(((((((..((.....((((...(((.....)))....)))))).)))))))......................---------------------------- ( -16.20, z-score = -0.88, R) >droVir3.scaffold_12963 6765077 93 + 20206255 -CCCUGCAGCUGUUGCCCACAACGUUACUGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCCACAGCCGC-CCGUACACUCUACACCUUGCG------------ -....(((((((..((.....((((....((.....)).....)))))).)))))))...........((...-..(((.....))).....)).------------ ( -16.20, z-score = 0.32, R) >droGri2.scaffold_15252 9955617 96 + 17193109 ----UGCAGCUGUUGCCCACAACGUUCCAGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCUGCCCCUAGC--CAUGCCCGCGACUCCUCAAACCGUGCA----- ----.(((((((..((.....((((...(((.....)))....)))))).)))))))..(((..((.......--...))..))).................----- ( -19.80, z-score = 0.30, R) >consensus UCUUUGCAGCUGUUGCCCACAACGUUUCGGUUCAACGCUUUUUACGUGCUCAGCUGCCACGCCCCUCCCCAUC___CCCCAUUCCAC_CUUUACA____________ .....(((((((..((.....((((...(((.....)))....)))))).))))))).................................................. (-15.32 = -15.42 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:22 2011