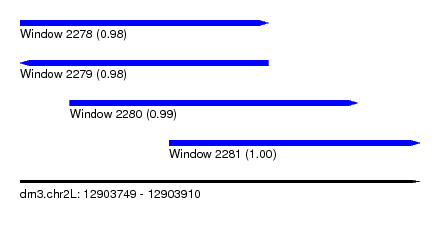
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,903,749 – 12,903,910 |
| Length | 161 |
| Max. P | 0.997996 |

| Location | 12,903,749 – 12,903,849 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.65220 |
| G+C content | 0.38491 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983284 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
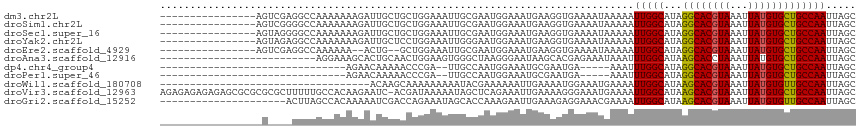
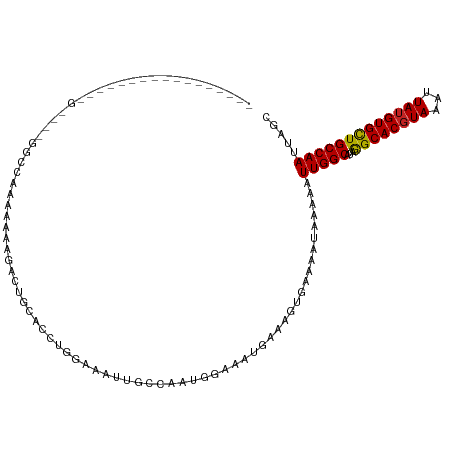
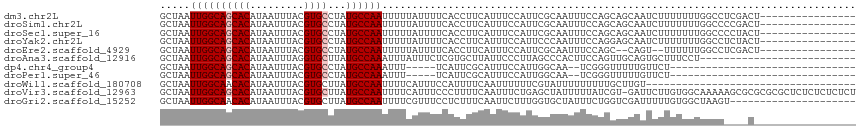
>dm3.chr2L 12903749 100 + 23011544 ----------------AGUCGAGGCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ----------------.......(((.........((((...........))))((....))...))).........((((((((...(((((((...)))))))))))))))... ( -20.90, z-score = -0.66, R) >droSim1.chr2L 12698026 100 + 22036055 ----------------AGUCGGGGCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ----------------..(((...(((.....((....))((........))...)))...))).............((((((((...(((((((...)))))))))))))))... ( -21.20, z-score = -0.55, R) >droSec1.super_16 1064133 100 + 1878335 ----------------AGUAGGGGCCAAAAAAAGAUUGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ----------------.((((...(((.....((....))..)))...)))).........................((((((((...(((((((...)))))))))))))))... ( -20.90, z-score = -0.69, R) >droYak2.chr2L 9324876 100 + 22324452 ----------------AGUAGAGGCCAAAAAAAGAUUGCUCCUGGAAAUUGGGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ----------------.......(((.............(((..(...)..)))((....))...))).........((((((((...(((((((...)))))))))))))))... ( -22.20, z-score = -1.54, R) >droEre2.scaffold_4929 14101596 96 - 26641161 ----------------AGUCGAGGCCAAAAAA--ACUG--GCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ----------------..((..(((((.....--..))--))).))..(..(..((....))....)..).......((((((((...(((((((...)))))))))))))))... ( -23.30, z-score = -1.98, R) >droAna3.scaffold_12916 3652315 90 - 16180835 --------------------------AGGAAAGCACUGCAACUGGAAGUGGGCUAAGGGAAUAAGCACGAGAAAUAAAUUUGGCAUAAGCACCUAAAUUAUGUGCUGCCAAUUAGC --------------------------.......((((.........)))).(((((.((....(((((((....).(((((((.........))))))).)))))).))..))))) ( -16.60, z-score = 0.87, R) >dp4.chr4_group4 560567 78 + 6586962 -------------------------------AGAACAAAAACCCGA--UUGCCAAUGGAAAUGCGAAUGA-----AAAUUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC -------------------------------............((.--((((..((....)))))).)).-----....((((((...(((((((...)))))))))))))..... ( -16.30, z-score = -0.84, R) >droPer1.super_46 95745 78 - 590583 -------------------------------AGAACAAAAACCCGA--UUGCCAAUGGAAAUGCGAAUGA-----AAAUUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC -------------------------------............((.--((((..((....)))))).)).-----....((((((...(((((((...)))))))))))))..... ( -16.30, z-score = -0.84, R) >droWil1.scaffold_180708 5943761 81 + 12563649 -----------------------------------ACAAGCAAAAAAAAAUACGAAAAAAUUGAAAAUGGAAAUGAAAAUUGGCAUAAGCACGUAAAUUAUGUGUUGCCAAUUAGC -----------------------------------..........................................((((((((...(((((((...)))))))))))))))... ( -14.80, z-score = -2.11, R) >droVir3.scaffold_12963 6761176 115 + 20206255 AGAGAGAGAGAGCGCGCGCGCUUUUUGCCACAAGAAUC-ACGAUAAAAAUAGCUCAGAAAUUGAAAAGGGAAAUGAAAAUUGGCAUAAGCACGUAAAUUAUGUGCUGCCAAUUAGC ...(..((((((((....))))))))..).........-............(((((....((....)).....))).((((((((...(((((((...))))))))))))))).)) ( -26.50, z-score = -0.90, R) >droGri2.scaffold_15252 9951536 95 + 17193109 ---------------------ACUUAGCCACAAAAAUCGACCAGAAAUAGCACCAAAGAAUUGAAAGAGGAAACGAAAAUUGGCAUAAGCACGUAAAUUAUGUGUUGCCAAUUAGC ---------------------............................((.................(....)...((((((((...(((((((...))))))))))))))).)) ( -17.40, z-score = -1.95, R) >consensus _________________G____GGCCAAAAAAAGACUGCACCUGGAAAUUGCCAAUGGAAAUGAAAGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGC ...............................................................................(((((...((((((((...)))))))))))))..... (-13.67 = -13.38 + -0.29)
| Location | 12,903,749 – 12,903,849 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.65220 |
| G+C content | 0.38491 |
| Mean single sequence MFE | -16.29 |
| Consensus MFE | -10.45 |
| Energy contribution | -10.64 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.978993 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 12903749 100 - 23011544 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUCGCAAUUUCCAGCAGCAAUCUUUUUUUGGCCUCGACU---------------- (((((((((((((((.........))))...)))))))).........................((........))))).....................---------------- ( -17.10, z-score = -1.67, R) >droSim1.chr2L 12698026 100 - 22036055 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUCGCAAUUUCCAGCAGCAAUCUUUUUUUGGCCCCGACU---------------- (((((((((((((((.........))))...)))))))).........................((........))))).........(((....)))..---------------- ( -17.30, z-score = -1.74, R) >droSec1.super_16 1064133 100 - 1878335 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUCGCAAUUUCCAGCAGCAAUCUUUUUUUGGCCCCUACU---------------- (((((((((((((((.........))))...)))))))).........................((........))))).....................---------------- ( -17.10, z-score = -2.07, R) >droYak2.chr2L 9324876 100 - 22324452 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUCCCAAUUUCCAGGAGCAAUCUUUUUUUGGCCUCUACU---------------- ...((((((((((((.........))))...)))))))).........................((((.....((((....))))...))))........---------------- ( -17.80, z-score = -2.25, R) >droEre2.scaffold_4929 14101596 96 + 26641161 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUCGCAAUUUCCAGC--CAGU--UUUUUUGGCCUCGACU---------------- ...((((((((((((.........))))...))))))))...................................((--(((.--....))))).......---------------- ( -18.60, z-score = -2.60, R) >droAna3.scaffold_12916 3652315 90 + 16180835 GCUAAUUGGCAGCACAUAAUUUAGGUGCUUAUGCCAAAUUUAUUUCUCGUGCUUAUUCCCUUAGCCCACUUCCAGUUGCAGUGCUUUCCU-------------------------- ((...((((((((((.........))))...))))))...........(.(((.........))).)..........))...........-------------------------- ( -14.50, z-score = 0.56, R) >dp4.chr4_group4 560567 78 - 6586962 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAAUUU-----UCAUUCGCAUUUCCAUUGGCAA--UCGGGUUUUUGUUCU------------------------------- .....((((((((((.........))))...))))))....-----..(((((.(((.((...)).))--)))))).........------------------------------- ( -14.30, z-score = -0.10, R) >droPer1.super_46 95745 78 + 590583 GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAAUUU-----UCAUUCGCAUUUCCAUUGGCAA--UCGGGUUUUUGUUCU------------------------------- .....((((((((((.........))))...))))))....-----..(((((.(((.((...)).))--)))))).........------------------------------- ( -14.30, z-score = -0.10, R) >droWil1.scaffold_180708 5943761 81 - 12563649 GCUAAUUGGCAACACAUAAUUUACGUGCUUAUGCCAAUUUUCAUUUCCAUUUUCAAUUUUUUCGUAUUUUUUUUUGCUUGU----------------------------------- (..((((((((.(((.........)))....))))))))..).....................(((........)))....----------------------------------- ( -10.30, z-score = -1.07, R) >droVir3.scaffold_12963 6761176 115 - 20206255 GCUAAUUGGCAGCACAUAAUUUACGUGCUUAUGCCAAUUUUCAUUUCCCUUUUCAAUUUCUGAGCUAUUUUUAUCGU-GAUUCUUGUGGCAAAAAGCGCGCGCGCUCUCUCUCUCU (..((((((((((((.........))))...))))))))..)...................((((..(((((..((.-.(...)..))..)))))((....))))))......... ( -23.70, z-score = -0.93, R) >droGri2.scaffold_15252 9951536 95 - 17193109 GCUAAUUGGCAACACAUAAUUUACGUGCUUAUGCCAAUUUUCGUUUCCUCUUUCAAUUCUUUGGUGCUAUUUCUGGUCGAUUUUUGUGGCUAAGU--------------------- (((....)))................(((((.((((....(((...((....((((....))))..........)).)))......)))))))))--------------------- ( -14.24, z-score = 0.29, R) >consensus GCUAAUUGGCAGCACAUAAUUUACGUGCCUAUGCCAAUUUUUAUUUUCACCUUCAUUUCCAUUGGCAAUUUCCAGCUGCAAUCUUUUUUUGGCC____C_________________ .....((((((((((.........))))...))))))............................................................................... (-10.45 = -10.64 + 0.18)

| Location | 12,903,769 – 12,903,885 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.14 |
| Shannon entropy | 0.58184 |
| G+C content | 0.40299 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
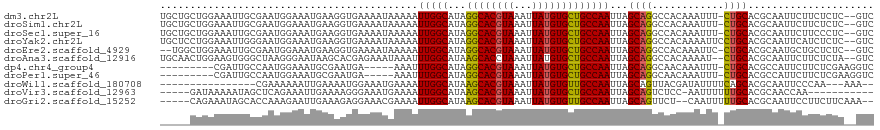
>dm3.chr2L 12903769 116 + 23011544 UGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUU-CUGCACGCAAUUCUUCUCUC--GUC .......((((((((((..(((((((....(((........((((((((...(((((((...))))))))))))))).......)))..))))-)))..))))))).)))....--... ( -31.46, z-score = -1.48, R) >droSim1.chr2L 12698046 116 + 22036055 UGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUU-CUGCACGCAAUUCUUCUCUC--GUC .......((((((((((..(((((((....(((........((((((((...(((((((...))))))))))))))).......)))..))))-)))..))))))).)))....--... ( -31.46, z-score = -1.48, R) >droSec1.super_16 1064153 116 + 1878335 UGCUGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUU-CUGCACGCAAUUCUUCCCUC--GUC .......((((((((((..(((((((....(((........((((((((...(((((((...))))))))))))))).......)))..))))-)))..))))))..))))...--... ( -33.06, z-score = -1.72, R) >droYak2.chr2L 9324896 117 + 22324452 UGCUCCUGGAAAUUGGGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAAUUCCUGCACGCAAUUCAUCUCUC--GUC ...(((..(...)..)))......(((((((((((......((((((((...(((((((...))))))))))))))).(((((..........)))))......))))))).))--)). ( -35.00, z-score = -2.61, R) >droEre2.scaffold_4929 14101614 114 - 26641161 --UGGCUGGAAAUUGCGAAUGGAAAUGAAGGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUC-CUGCACGCAAUGCUGCUCUC--GUC --.(((.....((((((........................((((((((...(((((((...))))))))))))))).(((((.........)-)))).))))))...)))...--... ( -33.10, z-score = -1.49, R) >droAna3.scaffold_12916 3652325 115 - 16180835 UGCAACUGGAAGUGGGCUAAGGGAAUAAGCACGAGAAAUAAAUUUGGCAUAAGCACCUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAAU--CUGCACGCAAUUCUUCUCUA--GUC ..............(((((.(((((.((...((..........((((((...((((.........))))))))))...(((((........)--)))).))...)).)))))))--))) ( -25.60, z-score = 0.33, R) >dp4.chr4_group4 560579 104 + 6586962 ---------CGAUUGCCAAUGGAAAUGCGAAUGA-----AAAUUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAAUUU-CUGCACGCCAUUCUUCUCGAAGGUC ---------.((((......((((..(((.....-----....((((((...(((((((...)))))))))))))...(((((.........)-)))).)))..)))).......)))) ( -26.02, z-score = -0.51, R) >droPer1.super_46 95757 104 - 590583 ---------CGAUUGCCAAUGGAAAUGCGAAUGA-----AAAUUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAAUUU-CUGCACGCCAUUCUUCUCGAAGGUC ---------.((((......((((..(((.....-----....((((((...(((((((...)))))))))))))...(((((.........)-)))).)))..)))).......)))) ( -26.02, z-score = -0.51, R) >droWil1.scaffold_180708 5943778 98 + 12563649 ----------------CGAAAAAAUUGAAAAUGGAAAUGAAAAUUGGCAUAAGCACGUAAAUUAUGUGUUGCCAAUUAGCAGUUACGAUAUUUUCAGCACGCAAUUCCCAA---AAA-- ----------------........(((((((((.((.((..((((((((...(((((((...)))))))))))))))..)).))....)))))))))..............---...-- ( -22.60, z-score = -2.50, R) >droVir3.scaffold_12963 6761216 102 + 20206255 -----GAUAAAAAUAGCUCAGAAAUUGAAAAGGGAAAUGAAAAUUGGCAUAAGCACGUAAAUUAUGUGCUGCCAAUUAGCAGUCUCC-AAUUUUUUGCACGCAACCAA----------- -----..........((..((((((((....((((..((..((((((((...(((((((...)))))))))))))))..)).)))))-))))))).))..........----------- ( -26.90, z-score = -2.90, R) >droGri2.scaffold_15252 9951556 110 + 17193109 -----CAGAAAUAGCACCAAAGAAUUGAAAGAGGAAACGAAAAUUGGCAUAAGCACGUAAAUUAUGUGUUGCCAAUUAGCAGUUCU--CAAUUUUUGCACGCAAUUCCUUCUUCAAA-- -----...................(((((..(((((.((..((((((((...(((((((...))))))))))))))).((((....--......)))).))...)))))..))))).-- ( -29.20, z-score = -3.49, R) >consensus _____CUGGAAAUUGCCAAUGGAAAUGAAAGUGAAAAUAAAAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAUUU_CUGCACGCAAUUCUUCUCUC__GUC ...........................................(((((...((((((((...)))))))))))))...((((............))))..................... (-16.92 = -16.57 + -0.35)
| Location | 12,903,809 – 12,903,910 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Shannon entropy | 0.41067 |
| G+C content | 0.40274 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.997996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
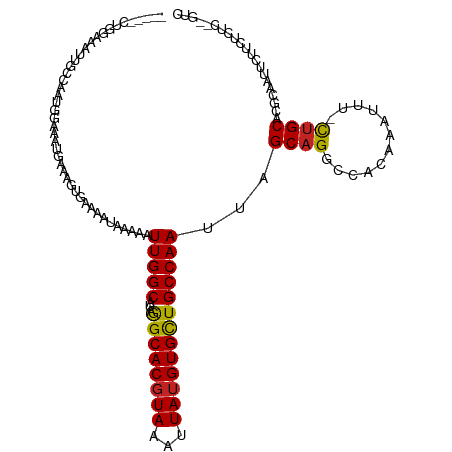
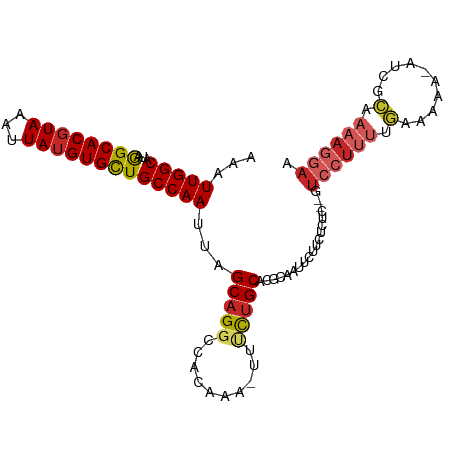
>dm3.chr2L 12903809 101 + 23011544 AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA-UUUCUGCACGCAAUUCUUCUCUC--GUCCUUUUGAAAAAACUCGCAAAAGGAA .((((((((...(((((((...))))))))))))))).(((((.......-..)))))................--.((((((((..........)))))))). ( -28.90, z-score = -3.13, R) >droSim1.chr2L 12698086 100 + 22036055 AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA-UUUCUGCACGCAAUUCUUCUCUC--GUCCUUUUGGAAAA-CUCGCAAAAGGAA .((((((((...(((((((...))))))))))))))).(((((.......-..)))))................--.((((((((((...-.)).)))))))). ( -30.90, z-score = -3.43, R) >droSec1.super_16 1064193 100 + 1878335 AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA-UUUCUGCACGCAAUUCUUCCCUC--GUCCUUUUGGAAAA-CUCGCAAAAGGAA .((((((((...(((((((...))))))))))))))).(((((.......-..)))))................--.((((((((((...-.)).)))))))). ( -30.90, z-score = -3.12, R) >droYak2.chr2L 9324936 101 + 22324452 AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAAAUUCCUGCACGCAAUUCAUCUCUC--GUCCUUUUGUGAAA-AUCGCAAAAGGAA .((((((((...(((((((...))))))))))))))).(((((..........)))))................--.((((((((((...-..)))))))))). ( -36.50, z-score = -5.31, R) >droEre2.scaffold_4929 14101652 100 - 26641161 AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA-UUCCUGCACGCAAUGCUGCUCUC--GUCCUUUUGCGAAA-AUCGCAAAAGGAA .((((((((...(((((((...))))))))))))))).(((((.......-..)))))..(((....)))....--.((((((((((...-..)))))))))). ( -40.40, z-score = -5.21, R) >droAna3.scaffold_12916 3652365 98 - 16180835 AAUUUGGCAUAAGCACCUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA--AUCUGCACGCAAUUCUUCUCUA--GUCCUUUGGAGAAA--UCACAAAAGGAA ...((((((...((((.........))))))))))...(((((.......--.)))))................--.((((((.((....--))...)))))). ( -24.80, z-score = -1.92, R) >dp4.chr4_group4 560608 98 + 6586962 ---UUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAA-UUUCUGCACGCCAUUCUUCUCGAAGGUCCUUUAGAAAAA--UCGCAAAAGGAA ---((((((...(((((((...)))))))))))))...(((((.......-..)))))...((.(((.....)))))((((((.((....--))...)))))). ( -25.00, z-score = -1.33, R) >droPer1.super_46 95786 98 - 590583 ---UUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCAACAAA-UUUCUGCACGCCAUUCUUCUCGAAGGUCCUUUAGAAAAA--UCGCAAAAGGAA ---((((((...(((((((...)))))))))))))...(((((.......-..)))))...((.(((.....)))))((((((.((....--))...)))))). ( -25.00, z-score = -1.33, R) >droVir3.scaffold_12963 6761251 85 + 20206255 AAAUUGGCAUAAGCACGUAAAUUAUGUGCUGCCAAUUAGCAGUCUCCAAU-UUUUUGCACGCAA--------------CCAAAAAAGAAGAAAAAUCAAA---- .((((((((...(((((((...))))))))))))))).....(((....(-((((((.......--------------.)))))))..))).........---- ( -19.60, z-score = -2.58, R) >droGri2.scaffold_15252 9951591 94 + 17193109 AAAUUGGCAUAAGCACGUAAAUUAUGUGUUGCCAAUUAGCAGUUCUCAAU-UUUU-GCACGCAAUUCCU--------UCUUCAAACGAAGUAGAAAAAAAUAAG .((((((((...(((((((...))))))))))))))).((((........-..))-)).........((--------.((((....)))).))........... ( -21.10, z-score = -2.86, R) >consensus AAAUUGGCAUAGGCACGUAAAUUAUGUGCUGCCAAUUAGCAGGCCACAAA_UUUCUGCACGCAAUUCUUCUCUC__GUCCUUUUGAAAAA_AUCGCAAAAGGAA ...(((((...((((((((...)))))))))))))...(((((..........)))))...................((((((.(..........).)))))). (-18.64 = -19.32 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:20 2011