
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,900,461 – 12,900,552 |
| Length | 91 |
| Max. P | 0.525997 |

| Location | 12,900,461 – 12,900,552 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.12 |
| Shannon entropy | 0.54925 |
| G+C content | 0.32399 |
| Mean single sequence MFE | -18.31 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525997 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
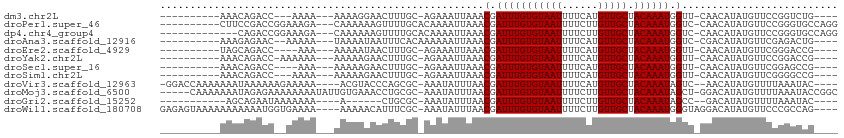
>dm3.chr2L 12900461 91 - 23011544 ----------AAACAGACC---AAAA---AAAAGGAACUUUGC-AGAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU-CAACAUAUGUUCCGGUCUG---- ----------...((((((---....---....(((((.((((-(.(((((....))))).))))).....((((((..((.....)).-))))))..)))))))))))---- ( -24.20, z-score = -3.46, R) >droPer1.super_46 91938 99 + 590583 ----------CUUCCGACCGGAAAGA---CAAAAAAGUUUUGCACAAAAUUAAACGAUUUGUGUAAUUUUCUUGUUGCUACAAAUGGUC-CAACAUAUGUUCCGGGUGCCAGG ----------.......((((((.((---(((.(((((..((((((((.........))))))))))))).)))))....((.(((...-...))).))))))))........ ( -22.60, z-score = -0.97, R) >dp4.chr4_group4 557069 96 - 6586962 -------------CAGACCGGAAAGA---CAAAAAAGUUUUGCACAAAAUUAAACGAUUUGUGUAAUUUUCUUGUUGCUACAAAUGGUC-CAACAUAUGUUCCGGGUGCCAGG -------------....((((((.((---(((.(((((..((((((((.........))))))))))))).)))))....((.(((...-...))).))))))))........ ( -22.60, z-score = -1.18, R) >droAna3.scaffold_12916 3648995 93 + 16180835 ----------AAAGAGAAC--AAAAA---UAAAAUAAUUUCACAAAAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUC-CGACAUAUGUUCGAGACUG---- ----------.....((((--(....---.....((((((......)))))).((.(((((((((((......))))).)))))).)).-.......))))).......---- ( -15.80, z-score = -1.38, R) >droEre2.scaffold_4929 14098298 90 + 26641161 ----------UAGCAGACC----AAA---AAAAAUAACUUUGC-AGAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU-CAACAUAUGUUCGGGACCG---- ----------..(((((..----...---.........)))))-............(((((((((((......))))).))))))((((-(.((....))...))))).---- ( -16.04, z-score = -0.76, R) >droYak2.chr2L 9321492 93 - 22324452 ----------AAACAGACC-AAAAAA---AAAAAGAACUUUGC-AGAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU-CAACAUAUGUUCCGGACCG---- ----------.......((-......---.....(((..((((-(.(((((....))))).)))))..)))((((((..((.....)).-)))))).......))....---- ( -16.70, z-score = -1.20, R) >droSec1.super_16 1060916 90 - 1878335 ----------AAACAGACC----AAA---AAAAAGAACUUUGC-AGAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU-CAACAUAUGUUCGGAGCCG---- ----------.......((----...---.....(((..((((-(.(((((....))))).)))))..)))((((((..((.....)).-))))))......)).....---- ( -16.70, z-score = -1.00, R) >droSim1.chr2L 12693193 91 - 22036055 ----------AAACAGACC---AAAA---AAAAAGAACUUUGC-AGAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU-CAACAUAUGUUCGGGGCCG---- ----------.......((---....---.....(((..((((-(.(((((....))))).)))))..)))((((((..((.....)).-))))))......)).....---- ( -16.90, z-score = -0.73, R) >droVir3.scaffold_12963 6758158 101 - 20206255 -GGACCAAAAAAAUAAAAAAGAAAAA----ACGUACCCAGCGC-AAAUAUUUAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUAGUC--AACAUAUGUUUUAAAUAC---- -......................(((----(((((....((((-((((........))))))))........((((((((....))).)--))))))))))))......---- ( -17.30, z-score = -2.12, R) >droMoj3.scaffold_6500 25941339 106 - 32352404 -----CAAAAAAAUAGAGAAAAAAAAUAUUGUGAAACCUGCGC-AAAUAUUUAACGAUUUGUGUAAUUUUCUUGUUGCUACAAAUAGCU-GGACAUAUGUUUUAAAUACCGGC -----.................((((((((((((((..(((((-((((........)))))))))..)))).....((((....)))).-..))).))))))).......... ( -17.10, z-score = -0.62, R) >droGri2.scaffold_15252 9948365 85 - 17193109 -----------AGCAGAAUAAAAAAA----A------CUGCGC-AAAUAUUUAACGAUUUGUGUAAUUUUCUUGUUGCUACAAAUAGCC--GACAUAUGUUUUAAAUAC---- -----------...((((((...(((----(------.(((((-((((........))))))))).))))..((((((((....))).)--))))..))))))......---- ( -14.80, z-score = -0.89, R) >droWil1.scaffold_180708 5940930 104 - 12563649 GAGAGUAAAAAAAAAAAUGGUGAAAA----AAAAACAUUUCGC-AAAUAUUUAACGAUUUGUGUAAUUUUCUUGUUGCUACAAAUGGGUAGGACAUAUGUUCCCGCCAG---- ...................((((((.----.......))))))-............(((((((((((......))))).)))))).((..((((....).)))..))..---- ( -19.00, z-score = -1.01, R) >consensus __________AAACAGACC__AAAAA___AAAAAAAACUUUGC_AAAAAUUAAACGAUUUGUGUAAUUUUCAUGUUGCUACAAAUGGUU_CAACAUAUGUUCCGGACAG____ ......................................................(.(((((((((((......))))).)))))).).......................... ( -8.30 = -8.30 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:16 2011