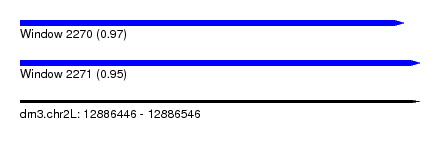
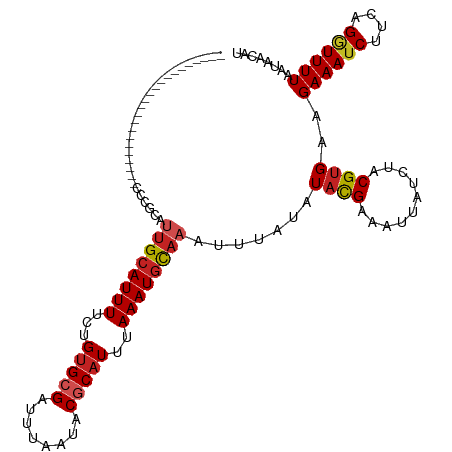
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,886,446 – 12,886,546 |
| Length | 100 |
| Max. P | 0.968552 |

| Location | 12,886,446 – 12,886,542 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Shannon entropy | 0.27458 |
| G+C content | 0.30032 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968552 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 12886446 96 + 23011544 -----------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAU -----------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))............ ( -22.70, z-score = -3.01, R) >droSim1.chr2L 12680522 96 + 22036055 -----------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAU -----------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))............ ( -22.70, z-score = -3.01, R) >droSec1.super_16 1048363 96 + 1878335 -----------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAU -----------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))............ ( -22.70, z-score = -3.01, R) >droYak2.chr2L 9308871 96 + 22324452 -----------------------UCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAU -----------------------......((((((((...(((((........)))))..)))))))).............((((...((.(((((....))))).))..))))..... ( -22.50, z-score = -2.95, R) >droEre2.scaffold_4929 14086079 96 - 26641161 -----------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGAUUUUAAUAACAU -----------------------..(((.((((((((...(((((........)))))..))))))))..........((.....))...)))..((((((....))))))........ ( -22.10, z-score = -3.14, R) >droAna3.scaffold_12916 3638164 96 - 16180835 -----------------------CCCUCAUGGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUUCGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAU -----------------------.....(((((((((...(((((........)))))..))))))...............((((...((.(((((....))))).))..))))..))) ( -21.50, z-score = -3.12, R) >dp4.chr4_group4 541771 96 + 6586962 -----------------------GUUUCGGUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAU -----------------------..((((.(((((((...(((((........)))))..)))))))..........))))((((...((.(((((....))))).))..))))..... ( -23.80, z-score = -3.39, R) >droPer1.super_46 76647 96 - 590583 -----------------------GUUUCGGUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAU -----------------------..((((.(((((((...(((((........)))))..)))))))..........))))((((...((.(((((....))))).))..))))..... ( -23.80, z-score = -3.39, R) >droWil1.scaffold_180708 5926918 96 + 12563649 -----------------------AUCGCAUUGCAUUUUCUGAGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAAAUAUGAAAUUAUCUACGUGAAGAAAUCUUCAGAUUUUAAUAACAU -----------------------.((((.((((((((.....(((........)))....)))))))).((((....)))).........)))).((((((....))))))........ ( -16.40, z-score = -0.92, R) >droMoj3.scaffold_6500 25925377 119 + 32352404 CCUUUCUGUGUAUGCGUGUGCAUCUCAUAUUGCAUUUUCUGUGCGAUUUAAUACCCAUUUAAAUGUAAAUUUGUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGCUUUUAAUAACAU .......((((...((((((((........(((((.....)))))((((((.......)))))).......))))))))............(((((....)))))..........)))) ( -20.30, z-score = -1.10, R) >droGri2.scaffold_15252 9936292 118 + 17193109 -AACGAAGUUUGUGUGCGUGUUCCUCUUAUUGCAUUUUCUGUGCGAUUUAAUACCCAUUUAAAUUCAAAUUUGCAUACGAAAUUAUCUACGUGAAGAAAUCUCUAGGUUUUAAUAAAAU -.(((.((((((((((((.(((.........((((.....))))(((((((.......)))))))..))).)))))))))).....)).)))...((((((....))))))........ ( -20.50, z-score = -1.07, R) >consensus _______________________CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAU .............................((((((((...(((((........)))))..)))))))).............((((....(.(((((....))))))....))))..... (-15.37 = -15.69 + 0.32)

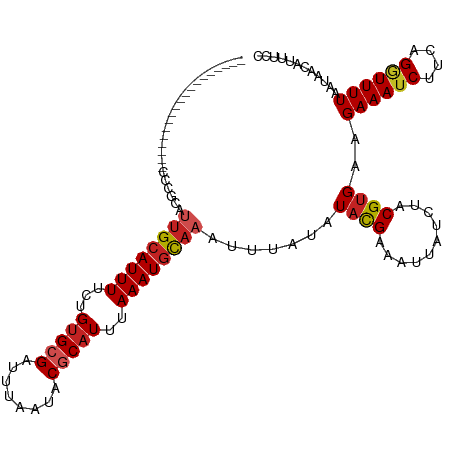
| Location | 12,886,446 – 12,886,546 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Shannon entropy | 0.26542 |
| G+C content | 0.30370 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12886446 100 + 23011544 -------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAUUUCC -------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))................ ( -22.70, z-score = -2.96, R) >droSim1.chr2L 12680522 100 + 22036055 -------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAUUUCC -------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))................ ( -22.70, z-score = -2.96, R) >droSec1.super_16 1048363 100 + 1878335 -------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAUUUCC -------------------(((...((((((((...(((((........)))))..))))))))........................((((....)))))))................ ( -22.70, z-score = -2.96, R) >droYak2.chr2L 9308871 100 + 22324452 -------------------UCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGGUUUUAAUAACAUUUCC -------------------......((((((((...(((((........)))))..)))))))).............((((...((.(((((....))))).))..))))......... ( -22.50, z-score = -2.89, R) >droEre2.scaffold_4929 14086079 100 - 26641161 -------------------CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCGGAUUUUAAUAACAUUUCC -------------------..(((.((((((((...(((((........)))))..))))))))..........((.....))...)))..((((((....))))))............ ( -22.10, z-score = -3.12, R) >droAna3.scaffold_12916 3638164 100 - 16180835 -------------------CCCUCAUGGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUUCGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAUUUAC -------------------.....(((((((((...(((((........)))))..))))))...............((((...((.(((((....))))).))..))))..))).... ( -21.60, z-score = -3.15, R) >dp4.chr4_group4 541771 100 + 6586962 -------------------GUUUCGGUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAUUUGC -------------------..((((.(((((((...(((((........)))))..)))))))..........))))((((...((.(((((....))))).))..))))......... ( -23.80, z-score = -2.95, R) >droPer1.super_46 76647 100 - 590583 -------------------GUUUCGGUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAUUUGC -------------------..((((.(((((((...(((((........)))))..)))))))..........))))((((...((.(((((....))))).))..))))......... ( -23.80, z-score = -2.95, R) >droWil1.scaffold_180708 5926918 100 + 12563649 -------------------AUCGCAUUGCAUUUUCUGAGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAAAUAUGAAAUUAUCUACGUGAAGAAAUCUUCAGAUUUUAAUAACAUUCAC -------------------.((((.((((((((.....(((........)))....)))))))).((((....)))).........)))).((((((....))))))............ ( -16.40, z-score = -0.87, R) >droMoj3.scaffold_6500 25925381 119 + 32352404 UCUGUGUAUGCGUGUGCAUCUCAUAUUGCAUUUUCUGUGCGAUUUAAUACCCAUUUAAAUGUAAAUUUGUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGCUUUUAAUAACAUUUAC ...((((...((((((((........(((((.....)))))((((((.......)))))).......))))))))............(((((....)))))..........)))).... ( -20.40, z-score = -0.84, R) >droGri2.scaffold_15252 9936296 118 + 17193109 -AAGUUUGUGUGCGUGUUCCUCUUAUUGCAUUUUCUGUGCGAUUUAAUACCCAUUUAAAUUCAAAUUUGCAUACGAAAUUAUCUACGUGAAGAAAUCUCUAGGUUUUAAUAAAAUUUAU -...((((((((((.(((.........((((.....))))(((((((.......)))))))..))).))))))))))..............((((((....))))))............ ( -19.40, z-score = -1.09, R) >consensus ___________________CCCGCAUUGCAUUUUCUGUGCGAUUUAAUACGCAUUUAAAUGCAAAUUUAUAUACGAAAUUAUCUACGUGAAGAAAUCUUCAGGUUUUAAUAACAUUUCC .........................((((((((...(((((........)))))..)))))))).............((((....(.(((((....))))))....))))......... (-15.37 = -15.69 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:12 2011