| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,871,417 – 12,871,513 |
| Length | 96 |
| Max. P | 0.647600 |

| Location | 12,871,417 – 12,871,513 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Shannon entropy | 0.44942 |
| G+C content | 0.37136 |
| Mean single sequence MFE | -11.20 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
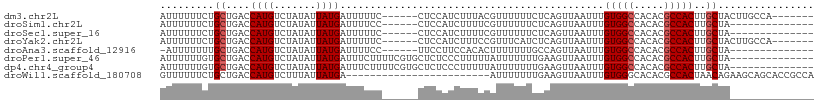
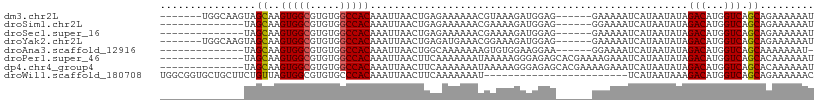
>dm3.chr2L 12871417 96 + 23011544 AUUUUUUCUGCUGACCAUGUCUAUAUUAUGAUUUUUC------CUCCAUCUUUACGUUUUUUCUCAGUUAAUUUGUGGCCACACGCCACUUGCUACUUGCCA------- .........(((((.((((.......)))).......------............(......))))))......(((((.....)))))..((.....))..------- ( -10.90, z-score = -0.79, R) >droSim1.chr2L 12665052 89 + 22036055 AUUUUUUCUGCUGACCAUGUCUAUAUUAUGAUUUUCC------CUCCAUCUUUUCGUUUUUUCUCAGUUAAUUUGUGGCCACACGCCACUUGCUA-------------- .........(((((.((((.......)))).......------............(......))))))......(((((.....)))))......-------------- ( -10.30, z-score = -1.31, R) >droSec1.super_16 1033261 89 + 1878335 AUUUUUUCUGCUGACCAUGUCUAUAUUAUGAUUUUUC------CUCCAUCUUUUCGUUUUUUCUCAGUUAAUUUGUGGCCACACGCCACUUGCUA-------------- .........(((((.((((.......)))).......------............(......))))))......(((((.....)))))......-------------- ( -10.30, z-score = -1.28, R) >droYak2.chr2L 9293823 96 + 22324452 AUUUUUUCUGCUGACCAUGUCUAUAUUAUGAUUUUUC------CUCCAUCUUUCCGUUUCAUCUCAGUUAAUUUGUGGCCACACGCCACUUGCUACUUGCCA------- .........(((((..(((..........(((.....------....))).........))).)))))......(((((.....)))))..((.....))..------- ( -11.11, z-score = -0.74, R) >droAna3.scaffold_12916 3622899 88 - 16180835 -AUUUUUUUGCUGACCAUGUCUAUAUUAUGAUUUUCC------UUCCUUCCACACUUUUUUUGCCAGUUAAUUUGUGGCCACACGCCACUUGCUA-------------- -........((....((((.......)))).......------...............................(((((.....)))))..))..-------------- ( -9.50, z-score = -0.63, R) >droPer1.super_46 61576 95 - 590583 AUUUUUUGUGCUGACCAUGUCUAUAUUAUGAUUUCUUUUCGUGCUCUCCCUUUUUAUUUUUUUGAAGUUAAUUUGUGGCCACACGCCACUUGCUA-------------- .........((.................(((((((............................)))))))....(((((.....)))))..))..-------------- ( -11.29, z-score = -0.51, R) >dp4.chr4_group4 528176 95 + 6586962 AUUUUUUGUGCUGACCAUGUCUAUAUUAUGAUUUCUUUUCGUGCUCUCCCUUUUUAUUUUUUUGAAGUUAAUUUGUGGCCACACGCCACUUGCUA-------------- .........((.................(((((((............................)))))))....(((((.....)))))..))..-------------- ( -11.29, z-score = -0.51, R) >droWil1.scaffold_180708 6647149 85 - 12563649 GUUUUUUCUGCUGACCAUGUCUUUAUUAUGA------------------------AUUUUUUUGAAGUUAAUUUGUGGGCACACGCCACUAACAGAAGCAGCACCGCCA ........(((((..((((.......)))).------------------------................(((((.(((....)))....)))))..)))))...... ( -14.90, z-score = 0.17, R) >consensus AUUUUUUCUGCUGACCAUGUCUAUAUUAUGAUUUUUC______CUCCAUCUUUUCGUUUUUUCUCAGUUAAUUUGUGGCCACACGCCACUUGCUA______________ .........((....((((.......))))............................................(((((.....)))))..))................ ( -8.14 = -8.39 + 0.25)
| Location | 12,871,417 – 12,871,513 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Shannon entropy | 0.44942 |
| G+C content | 0.37136 |
| Mean single sequence MFE | -13.61 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.29 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
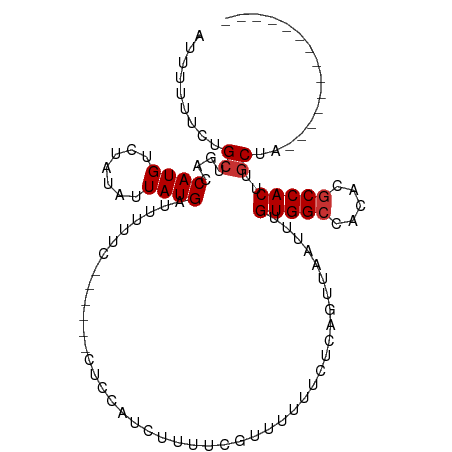
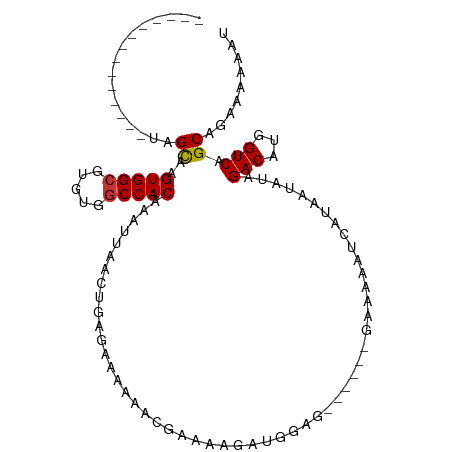
>dm3.chr2L 12871417 96 - 23011544 -------UGGCAAGUAGCAAGUGGCGUGUGGCCACAAAUUAACUGAGAAAAAACGUAAAGAUGGAG------GAAAAAUCAUAAUAUAGACAUGGUCAGCAGAAAAAAU -------..((.....))..(((((.....))))).......(((..............(((....------.....)))........(((...)))..)))....... ( -13.60, z-score = -0.20, R) >droSim1.chr2L 12665052 89 - 22036055 --------------UAGCAAGUGGCGUGUGGCCACAAAUUAACUGAGAAAAAACGAAAAGAUGGAG------GGAAAAUCAUAAUAUAGACAUGGUCAGCAGAAAAAAU --------------......(((((.....))))).......(((..............(((....------.....)))........(((...)))..)))....... ( -12.60, z-score = -0.96, R) >droSec1.super_16 1033261 89 - 1878335 --------------UAGCAAGUGGCGUGUGGCCACAAAUUAACUGAGAAAAAACGAAAAGAUGGAG------GAAAAAUCAUAAUAUAGACAUGGUCAGCAGAAAAAAU --------------......(((((.....))))).......(((..............(((....------.....)))........(((...)))..)))....... ( -12.60, z-score = -1.11, R) >droYak2.chr2L 9293823 96 - 22324452 -------UGGCAAGUAGCAAGUGGCGUGUGGCCACAAAUUAACUGAGAUGAAACGGAAAGAUGGAG------GAAAAAUCAUAAUAUAGACAUGGUCAGCAGAAAAAAU -------..((.....))..(((((.....))))).......(((...(((.........(((...------((....))....)))........))).)))....... ( -14.23, z-score = -0.21, R) >droAna3.scaffold_12916 3622899 88 + 16180835 --------------UAGCAAGUGGCGUGUGGCCACAAAUUAACUGGCAAAAAAAGUGUGGAAGGAA------GGAAAAUCAUAAUAUAGACAUGGUCAGCAAAAAAAU- --------------......(((((.....))))).......(((((.......((((.....((.------......))...)))).......))))).........- ( -14.34, z-score = -0.76, R) >droPer1.super_46 61576 95 + 590583 --------------UAGCAAGUGGCGUGUGGCCACAAAUUAACUUCAAAAAAAUAAAAAGGGAGAGCACGAAAAGAAAUCAUAAUAUAGACAUGGUCAGCACAAAAAAU --------------..((..(((((.....))))).......((((..............)))).)).....................(((...)))............ ( -13.34, z-score = -1.05, R) >dp4.chr4_group4 528176 95 - 6586962 --------------UAGCAAGUGGCGUGUGGCCACAAAUUAACUUCAAAAAAAUAAAAAGGGAGAGCACGAAAAGAAAUCAUAAUAUAGACAUGGUCAGCACAAAAAAU --------------..((..(((((.....))))).......((((..............)))).)).....................(((...)))............ ( -13.34, z-score = -1.05, R) >droWil1.scaffold_180708 6647149 85 + 12563649 UGGCGGUGCUGCUUCUGUUAGUGGCGUGUGCCCACAAAUUAACUUCAAAAAAAU------------------------UCAUAAUAAAGACAUGGUCAGCAGAAAAAAC ..(((....)))(((((((.((((.(....)))))...................------------------------..........(((...))))))))))..... ( -14.80, z-score = 0.54, R) >consensus ______________UAGCAAGUGGCGUGUGGCCACAAAUUAACUGAGAAAAAACGAAAAGAUGGAG______GAAAAAUCAUAAUAUAGACAUGGUCAGCAGAAAAAAU ................((..(((((.....))))).....................................................(((...))).))......... (-11.27 = -11.29 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:09 2011