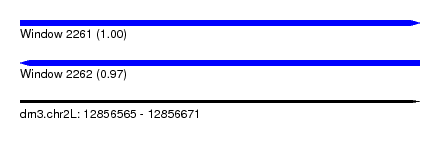
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,856,565 – 12,856,671 |
| Length | 106 |
| Max. P | 0.998978 |

| Location | 12,856,565 – 12,856,671 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.17885 |
| G+C content | 0.44064 |
| Mean single sequence MFE | -41.08 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.37 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.998978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
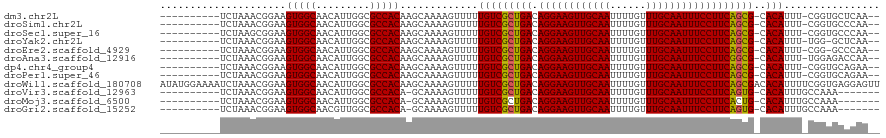
>dm3.chr2L 12856565 106 + 23011544 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUGCUCAA-- ----------..........((((....))))((((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.))))))...-- ( -42.10, z-score = -4.71, R) >droSim1.chr2L 12651577 106 + 22036055 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUGCCCAA-- ----------..........((((....))))((((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.))))))...-- ( -44.30, z-score = -5.18, R) >droSec1.super_16 1019856 106 + 1878335 ----------UCUAAGCGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUGCCCAA-- ----------..........((((....))))((((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.))))))...-- ( -44.30, z-score = -4.88, R) >droYak2.chr2L 9279601 105 + 22324452 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-UGG-GCUCAA-- ----------...........(((((.........))))).(((..(((....(((((((((.((((((((((((......))))))))))))))))))-.))).))-)..-)))...-- ( -39.80, z-score = -4.08, R) >droEre2.scaffold_4929 14057777 105 - 26641161 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGG-GCCCAA-- ----------.....((....))(....).((((.(((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-..)-))))))-- ( -41.00, z-score = -4.31, R) >droAna3.scaffold_12916 3610562 106 - 16180835 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCGGCG-CACAUUU-UGGAGACCAA-- ----------(((...((((((((((.........))))).............(((((((((.((((((((((((......))))))))))))))))))-.))))))-)).)))....-- ( -39.20, z-score = -3.71, R) >dp4.chr4_group4 510715 106 + 6586962 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUGCAGAA-- ----------(((.......((((....)))).(((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.)))))))).-- ( -42.60, z-score = -4.72, R) >droPer1.super_46 42587 106 - 590583 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG-CACAUUU-CGGUGCAGAA-- ----------(((.......((((....)))).(((((..((((....)))).(((((((((.((((((((((((......))))))))))))))))))-.)))...-.)))))))).-- ( -42.60, z-score = -4.72, R) >droWil1.scaffold_180708 5887834 120 + 12563649 AUAUGGAAAAUCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCGACACAUUUUCGGUGAGGAGUU ..........(((..((.((((((((.........))))).............(((((((((.((((((((((((......)))))))))))))))))))))....))).))..)))... ( -46.40, z-score = -5.32, R) >droVir3.scaffold_12963 6709520 101 + 20206255 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGUG-CACAUUUGCCAAA------- ----------.....((....))(....).(((((((....-)).........(((((((((.((((((((((((......))))))))))))))))))-.)))...))))).------- ( -38.10, z-score = -4.00, R) >droMoj3.scaffold_6500 25889989 101 + 32352404 ----------UCUAAACGGAAGUGGCAACAUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCACUG-CACAUUUGCCAAA------- ----------.....((....))(((((...((((((.(((-(.........)))).))(((.((((((((((((......)))))))))))))))..)-).)).)))))...------- ( -32.40, z-score = -2.39, R) >droGri2.scaffold_15252 9908623 101 + 17193109 ----------UCUAAACGGAAGUGGCAACGUUGGCGCCACA-GCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGUG-CACAUUUGCCAAA------- ----------.....((....))(((((.((((......))-)).........(((((((((.((((((((((((......))))))))))))))))))-.))).)))))...------- ( -40.10, z-score = -4.36, R) >consensus __________UCUAAACGGAAGUGGCAACAUUGGCGCCACAAGCAAAAGUUUUUGUCGCUGACAGGAAGUUGCAAUUUUGUUUGCAAUUUCCUUCAGCG_CACAUUU_CGGUGCCCAA__ .....................(((((.........))))).............(((((((((.((((((((((((......))))))))))))))))))..)))................ (-33.86 = -33.68 + -0.18)
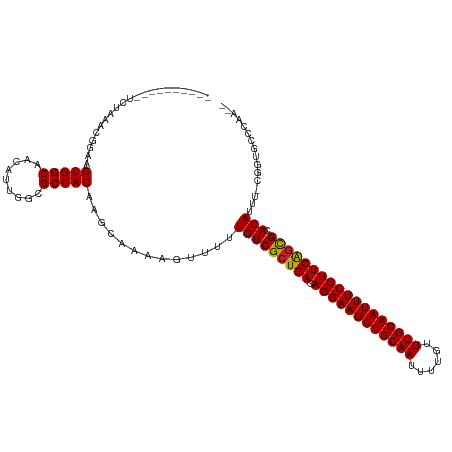
| Location | 12,856,565 – 12,856,671 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.17885 |
| G+C content | 0.44064 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12856565 106 - 23011544 --UUGAGCACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --..(((((...-.....((-(((((((((((.((((((......)))))).))))).)))))).))........)))))((((((.......))))))...........---------- ( -36.39, z-score = -3.61, R) >droSim1.chr2L 12651577 106 - 22036055 --UUGGGCACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --..(((((...-.....((-(((((((((((.((((((......)))))).))))).)))))).))........)))))((((((.......))))))...........---------- ( -35.59, z-score = -2.75, R) >droSec1.super_16 1019856 106 - 1878335 --UUGGGCACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGCUUAGA---------- --((((((....-...(((.-(((((((((((.((((((......)))))).))))).))))))))).............((((((.......))))))....)))))).---------- ( -37.50, z-score = -3.35, R) >droYak2.chr2L 9279601 105 - 22324452 --UUGAGC-CCA-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --((((((-...-...(((.-(((((((((((.((((((......)))))).))))).))))))))).............((((((.......))))))....)))))).---------- ( -35.00, z-score = -3.53, R) >droEre2.scaffold_4929 14057777 105 + 26641161 --UUGGGC-CCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --..(((.-...-...(((.-(((((((((((.((((((......)))))).))))).))))))))).............((((((.......)))))).))).......---------- ( -34.30, z-score = -2.56, R) >droAna3.scaffold_12916 3610562 106 + 16180835 --UUGGUCUCCA-AAAUGUG-CGCCGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --.((((...((-...((.(-(((((((((((.((((((......)))))).))))).)).((((((((....)))).))))))))))).))..))))............---------- ( -29.80, z-score = -1.71, R) >dp4.chr4_group4 510715 106 - 6586962 --UUCUGCACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --.((((.((.(-((.(((.-(((((((((((.((((((......)))))).))))).))))))))).............((((((.......))))))))).)).))))---------- ( -36.60, z-score = -3.96, R) >droPer1.super_46 42587 106 + 590583 --UUCUGCACCG-AAAUGUG-CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- --.((((.((.(-((.(((.-(((((((((((.((((((......)))))).))))).))))))))).............((((((.......))))))))).)).))))---------- ( -36.60, z-score = -3.96, R) >droWil1.scaffold_180708 5887834 120 - 12563649 AACUCCUCACCGAAAAUGUGUCGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGAUUUUCCAUAU ...........((((((.((((((((((((((.((((((......)))))).))))).)))))))))....((...((..((((((.......))))))....))..))))))))..... ( -39.60, z-score = -5.46, R) >droVir3.scaffold_12963 6709520 101 - 20206255 -------UUUGGCAAAUGUG-CACUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- -------...((((((.((.-(.(((((((((.((((((......)))))).))))).)))).)......)).)))))-)((((((.......))))))...........---------- ( -31.00, z-score = -2.26, R) >droMoj3.scaffold_6500 25889989 101 - 32352404 -------UUUGGCAAAUGUG-CAGUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAAUGUUGCCACUUCCGUUUAGA---------- -------..((((((..(((-(.....(((((.((((((......)))))).)))))..((((((.........))))-))..)))).....))))))............---------- ( -29.70, z-score = -1.51, R) >droGri2.scaffold_15252 9908623 101 - 17193109 -------UUUGGCAAAUGUG-CACUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGC-UGUGGCGCCAACGUUGCCACUUCCGUUUAGA---------- -------...((((((.((.-(.(((((((((.((((((......)))))).))))).)))).)......)).)))))-)((((((.......))))))...........---------- ( -31.00, z-score = -2.20, R) >consensus __UUGGGCACCG_AAAUGUG_CGCUGAAGGAAAUUGCAAACAAAAUUGCAACUUCCUGUCAGCGACAAAAACUUUUGCUUGUGGCGCCAAUGUUGCCACUUCCGUUUAGA__________ .............(((((((.(.(((((((((.((((((......)))))).))))).)))).).)).............((((((.......))))))...)))))............. (-24.37 = -24.62 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:05 2011