| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,204,207 – 1,204,300 |
| Length | 93 |
| Max. P | 0.539949 |

| Location | 1,204,207 – 1,204,300 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 65.05 |
| Shannon entropy | 0.64763 |
| G+C content | 0.55461 |
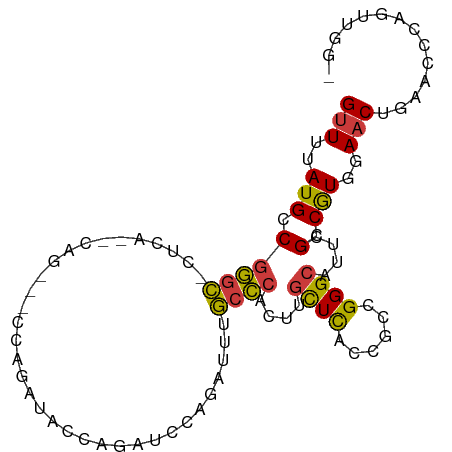
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -10.73 |
| Energy contribution | -10.12 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
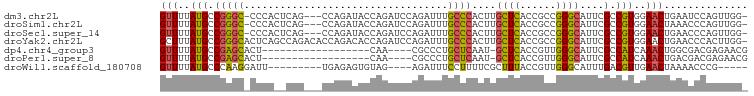
>dm3.chr2L 1204207 93 - 23011544 GUUUUAUGCCGGGC-CCCACUCAG---CCAGAUACCAGAUCCAGAUUUGCCCACUUGCUCACCGCCGGGCAUUCGCCGUGGAACUGAAUCCAGUUGG- ((...(((((.(((-.......((---(.((....(((((....)))))....)).)))....))).)))))..)).....(((((....)))))..- ( -22.20, z-score = 0.87, R) >droSim1.chr2L 1183506 93 - 22036055 GUUUUAUGCCGGGC-CCCACUCAG---CCAGAUACCAGAUCCAGAUUUGCCCACUUGCUCACCGCCGGGCAUUCGCCGUGGAACUAAACCCAGUUGG- ((((((((.((.((-((.....((---(.((....(((((....)))))....)).))).......))))...)).)))))))).............- ( -20.80, z-score = 0.96, R) >droSec1.super_14 1162725 93 - 2068291 GUUUUAUGCCGGGC-CCCACUCAG---CCAGAUACCAGAUCCAGAUUUGCCCACUUGCUCACCGCCGGGCAUUCGCCGUGGAACUGAACCCAGUUGG- ((...(((((.(((-.......((---(.((....(((((....)))))....)).)))....))).)))))..)).....(((((....)))))..- ( -22.20, z-score = 0.83, R) >droYak2.chr2L 1185441 97 - 22324452 GCUUUAUGCCGGGCACUCAGCCAGACACCAGACACCAGAUCCAGAUUUGCCCACUUGCUCACCGCCGGGCAUUCGCCGUGGAACUGAACCCACUUGG- ((...(((((.(((....(((.((.(....)....(((((....)))))....)).)))....))).)))))..)).((((........))))....- ( -24.30, z-score = -0.07, R) >dp4.chr4_group3 9664085 75 - 11692001 GUUUUAUGCCGAGCACU------------------CAA----CGCCCUGCUCAAU-GCUCACCGUUGGGCAUUCGCCAUCAAACUGGCGACGAGAACG (((((.....(((((..------------------...----.....))))).((-(((((....)))))))((((((......))))))..))))). ( -23.50, z-score = -2.17, R) >droPer1.super_8 862667 75 - 3966273 GUUUUAUGCCGAGCACU------------------CAA----CGCCCUGCUCAAU-GCUCACCGUUGGGCAUUCGCCAUCAAACUGACGACGAGAACG ((((.(((.((((..((------------------(((----((....((.....-))....)))))))..)))).))).)))).............. ( -19.50, z-score = -1.47, R) >droWil1.scaffold_180708 9751646 80 + 12563649 GUUUUAUGCCCAAGGAUU---------UGAGAGUGUAG----AGAUUUCCUUUUCGCUUUACCGUUGGGCAUUUGACGUUGAACUAAAACCCG----- (((..((((((((((...---------..((((((.((----((.....)))).)))))).)).))))))))..)))................----- ( -25.00, z-score = -3.84, R) >consensus GUUUUAUGCCGGGC_CUCA__CAG___CCAGAUACCAGAUCCAGAUUUGCCCACUUGCUCACCGCCGGGCAUUCGCCGUGGAACUGAACCCAGUUGG_ (((..(((.(((((..................................))))....((((......))))....).)))..))).............. (-10.73 = -10.12 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:52 2011