| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,847,267 – 12,847,373 |
| Length | 106 |
| Max. P | 0.569865 |

| Location | 12,847,267 – 12,847,373 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.24 |
| Shannon entropy | 0.16009 |
| G+C content | 0.38313 |
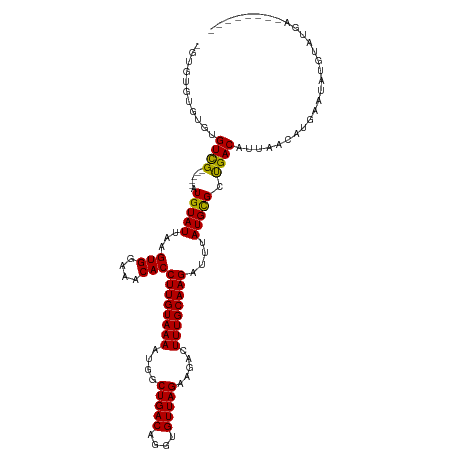
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.569865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
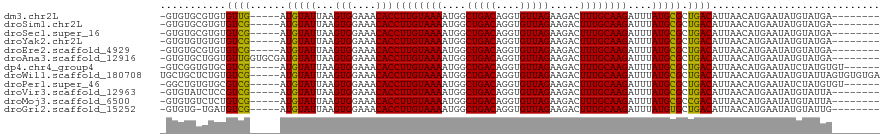
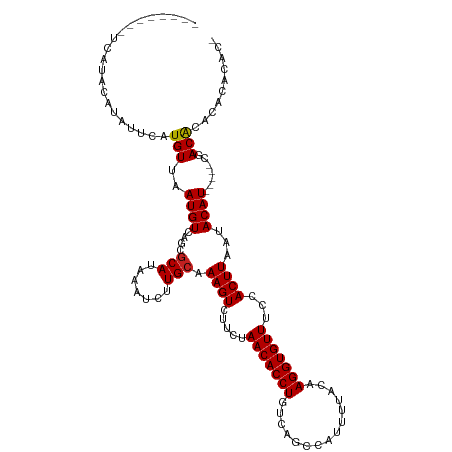
>dm3.chr2L 12847267 106 + 23011544 -GUGUGCGUGUGUUG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -..((((((((((((-----((((.....(((....)))((((((((....(((((....))))).....))))))))............))))))))))....))))))..-------- ( -31.00, z-score = -3.04, R) >droSim1.chr2L 12642314 106 + 22036055 -GUGUGCGUGUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -((((....((((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))))..))))............-------- ( -29.50, z-score = -2.21, R) >droSec1.super_16 1010727 106 + 1878335 -GUGUGCGUGUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -((((....((((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))))..))))............-------- ( -29.50, z-score = -2.21, R) >droYak2.chr2L 9270000 106 + 22324452 -GUGUGUGUGUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -..((((..((((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))))..))))............-------- ( -29.60, z-score = -2.50, R) >droEre2.scaffold_4929 14048491 106 - 26641161 -GUGUGCGUGUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -((((....((((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))))..))))............-------- ( -29.50, z-score = -2.21, R) >droAna3.scaffold_12916 3601522 111 - 16180835 -GUGUGCUGGUGUUGGUGCGAUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA-------- -..(((.((((((..(((((.........(((....)))((((((((....(((((....))))).....)))))))).....)))))..)))))).)))............-------- ( -35.40, z-score = -3.80, R) >dp4.chr4_group4 497476 108 + 6586962 -GUCGGUGUGCGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUCUAUGUGU------ -((((((((......-----.........(((....)))((((((((....(((((....))))).....))))))))......))))))))....(((((......)))))..------ ( -31.20, z-score = -2.26, R) >droWil1.scaffold_180708 5868762 115 + 12563649 UGCUGCUCUGUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUUAGUGUGUGA .((((....((((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))))..((((....))))..))))...... ( -28.10, z-score = -0.87, R) >droPer1.super_46 28122 108 - 590583 -GGCUGUGUGCGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUCUAUGUGU------ -.((.....))((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))....(((((......)))))..------ ( -28.20, z-score = -1.33, R) >droVir3.scaffold_12963 6697391 106 + 20206255 -GUGUAUCUCCGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUUA-------- -..........((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).))))....((((....))))....-------- ( -25.20, z-score = -1.25, R) >droMoj3.scaffold_6500 25877555 106 + 32352404 -GUGUGUCUCUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCCGACAUUAACAUGAAUAUGUAUUA-------- -..((((...(((((-----.(((((...(((....)))((((((((....(((((....))))).....))))))))....))))).)))))...))))............-------- ( -29.10, z-score = -2.32, R) >droGri2.scaffold_15252 9895511 105 + 17193109 -GUGUG-UGAUGUCG-----AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGUGCUGACAUUAACAUGAAUAUGUAUUG-------- -((((.-((((((((-----..((((...(((....)))((((((((....(((((....))))).....))))))))......))))))))))))))))............-------- ( -28.50, z-score = -2.70, R) >consensus _GUGUGUGUGUGUCG_____AUGUAUUAAGUGGAAACACCUUGUAAAAUGGCUGACAGGUGUUAGAAGACUUUGCAAGAUUUAUGCGCUGACAUUAACAUGAAUAUGUAUGA________ ...........(((...............(((....)))((((((((....(((((....))))).....))))))))...........)))............................ (-20.44 = -20.30 + -0.14)
| Location | 12,847,267 – 12,847,373 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.24 |
| Shannon entropy | 0.16009 |
| G+C content | 0.38313 |
| Mean single sequence MFE | -17.83 |
| Consensus MFE | -14.67 |
| Energy contribution | -14.79 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12847267 106 - 23011544 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CAACACACGCACAC- --------.............((((.((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----.)))).........- ( -16.06, z-score = -1.71, R) >droSim1.chr2L 12642314 106 - 22036055 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACACACGCACAC- --------.............(((..((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----..))).........- ( -17.36, z-score = -1.69, R) >droSec1.super_16 1010727 106 - 1878335 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACACACGCACAC- --------.............(((..((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----..))).........- ( -17.36, z-score = -1.69, R) >droYak2.chr2L 9270000 106 - 22324452 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACACACACACAC- --------.............(((..((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----..))).........- ( -17.36, z-score = -2.05, R) >droEre2.scaffold_4929 14048491 106 + 26641161 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACACACGCACAC- --------.............(((..((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----..))).........- ( -17.36, z-score = -1.69, R) >droAna3.scaffold_12916 3601522 111 + 16180835 --------UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAUCGCACCAACACCAGCACAC- --------.............((((..(((..(((((.......))).((((.....(((((((...............)))))))...))))........))....)))..))))...- ( -16.06, z-score = -1.33, R) >dp4.chr4_group4 497476 108 - 6586962 ------ACACAUAGAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACGCACACCGAC- ------.......((((((......)))))).(((((.......))).((((.....(((((((...............)))))))...)))).......-----....))........- ( -17.66, z-score = -0.91, R) >droWil1.scaffold_180708 5868762 115 - 12563649 UCACACACUAAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACACAGAGCAGCA .....................((((..((((...(((.......))).((((.....(((((((...............)))))))...)))).......-----.))))...))))... ( -18.96, z-score = -1.14, R) >droPer1.super_46 28122 108 + 590583 ------ACACAUAGAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACGCACACAGCC- ------.......((((((......)))))).(((((.......))).((((.....(((((((...............)))))))...)))).......-----....))........- ( -17.66, z-score = -0.91, R) >droVir3.scaffold_12963 6697391 106 - 20206255 --------UAAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACGGAGAUACAC- --------.......((((..(((..((((....(((.......))).((((.....(((((((...............)))))))...))))...))))-----..)))..))))...- ( -19.26, z-score = -1.49, R) >droMoj3.scaffold_6500 25877555 106 - 32352404 --------UAAUACAUAUUCAUGUUAAUGUCGGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACAGAGACACAC- --------.............((((..((((((.(((.......))).((((.....(((((((...............)))))))...))))......)-----)))))..))))...- ( -22.36, z-score = -2.61, R) >droGri2.scaffold_15252 9895511 105 - 17193109 --------CAAUACAUAUUCAUGUUAAUGUCAGCACAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU-----CGACAUCA-CACAC- --------............((((..((((..(((.........))).((((.....(((((((...............)))))))...))))...))))-----..))))..-.....- ( -16.56, z-score = -2.07, R) >consensus ________UCAUACAUAUUCAUGUUAAUGUCAGCGCAUAAAUCUUGCAAAGUCUUCUAACACCUGUCAGCCAUUUUACAAGGUGUUUCCACUUAAUACAU_____CGACACACACACAC_ .....................(((...((((...(((.......))).((((.....(((((((...............)))))))...)))).............))))...))).... (-14.67 = -14.79 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:36:01 2011