
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,835,967 – 12,836,096 |
| Length | 129 |
| Max. P | 0.920719 |

| Location | 12,835,967 – 12,836,070 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.23 |
| Shannon entropy | 0.66655 |
| G+C content | 0.30270 |
| Mean single sequence MFE | -18.11 |
| Consensus MFE | -6.58 |
| Energy contribution | -7.67 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12835967 103 + 23011544 AUUUAUUGUUAGUUUUUAUACUCGAUUAGAUCUCU--UUAAAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGC .....((((((((.....(((.(((((((....))--....))))).)))......(((((((.......((((......)))).....))))))))))))))). ( -20.60, z-score = -2.46, R) >droEre2.scaffold_4929 14037533 103 - 26641161 CUUUAUUUUGAGUUCUUGUACCCGAUUAGAUCCGU--GUAUAUGUGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGC ................(((((.((........)).--))))).....((.(((.(((((((((.......((((......)))).....))))))))).))).)) ( -19.50, z-score = -1.01, R) >droYak2.chr2L 9258592 105 + 22324452 AUUUAUUUUUAACUCUUGCACGCGAUUAGAUCUCUCUGUAAAUUUGCGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCGAUAUCCCAUUAAUUGAUAGC .................((((((((.((((....)))).....)))))).(((.(((((((((.......((((......)))).....))))))))).))).)) ( -24.40, z-score = -2.82, R) >droSec1.super_16 999501 103 + 1878335 AUUUAUUUUUAGUUUUUACACCCGAUUAGAUCUCU--GUAAAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGC ................(((((.....(((....))--).......)))))(((.(((((((((.......((((......)))).....))))))))).)))... ( -19.40, z-score = -2.03, R) >droSim1.chr2L 12630799 103 + 22036055 AUUUAUUGUUAGUUUUUACACCCGAUUAGAUCUCU--GUAAAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGC .....((((((((..((((((.....(((....))--).......)))))).....(((((((.......((((......)))).....))))))))))))))). ( -20.30, z-score = -2.00, R) >dp4.chr4_group4 485430 95 + 6586962 UUUUGUAUGCAGUUUAUCAAGAAAAGGAAAACU-------AAUUUAUAUAAC-UAUUUUUAGCCAUCAAAUAUUUACUUUUUCCACCAUUUUGGUUAGCCAGC-- ........((.(((.(((((((...((((((..-------(((........(-((....))).........)))....))))))....))))))).)))..))-- ( -9.03, z-score = 0.96, R) >droWil1.scaffold_180708 5855516 104 + 12563649 GUUUAAUCUUAACUAAACUAUUUGGCAACAAUUUACUUUAAUUUCGUGCAAAGUUUUAAAGAAUUUCUGUUCAGU-CAAAAAGCACAAUUCCAGAAAGUGAUAAC (((((........)))))...(((....))).(((((((......((((....(((.(..((((....))))..)-.)))..))))........))))))).... ( -13.54, z-score = -0.10, R) >consensus AUUUAUUUUUAGUUUUUACACCCGAUUAGAUCUCU__GUAAAUUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGC ......................................................(((((((((.......((((......)))).....)))))))))....... ( -6.58 = -7.67 + 1.09)



| Location | 12,835,967 – 12,836,070 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.23 |
| Shannon entropy | 0.66655 |
| G+C content | 0.30270 |
| Mean single sequence MFE | -18.69 |
| Consensus MFE | -7.18 |
| Energy contribution | -8.81 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12835967 103 - 23011544 GCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUUUAA--AGAGAUCUAAUCGAGUAUAAAAACUAACAAUAAAU .......(((((((((.....((((......)))).......)))))))))....(((.(((((....--........))))).))).................. ( -20.20, z-score = -3.14, R) >droEre2.scaffold_4929 14037533 103 + 26641161 GCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACACAUAUAC--ACGGAUCUAAUCGGGUACAAGAACUCAAAAUAAAG .......(((((((((.....((((......)))).......))))))))).................--............((((......))))......... ( -18.80, z-score = -2.26, R) >droYak2.chr2L 9258592 105 - 22324452 GCUAUCAAUUAAUGGGAUAUCGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACGCAAAUUUACAGAGAGAUCUAAUCGCGUGCAAGAGUUAAAAAUAAAU ((.....(((((((((.....((((......)))).......))))))))).....((((..(((...(((....)))))).))))))................. ( -20.00, z-score = -1.41, R) >droSec1.super_16 999501 103 - 1878335 GCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUUUAC--AGAGAUCUAAUCGGGUGUAAAAACUAAAAAUAAAU .......(((((((((.....((((......)))).......)))))))))...((((((((((((..--.)))))).......))))))............... ( -21.11, z-score = -2.69, R) >droSim1.chr2L 12630799 103 - 22036055 GCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUUUAC--AGAGAUCUAAUCGGGUGUAAAAACUAACAAUAAAU .......(((((((((.....((((......)))).......)))))))))...((((((((((((..--.)))))).......))))))............... ( -21.11, z-score = -2.60, R) >dp4.chr4_group4 485430 95 - 6586962 --GCUGGCUAACCAAAAUGGUGGAAAAAGUAAAUAUUUGAUGGCUAAAAAUA-GUUAUAUAAAUU-------AGUUUUCCUUUUCUUGAUAAACUGCAUACAAAA --((.(..(((((.....)))((((((.......((((((((((((....))-))))).))))).-------..)))))).........))..).))........ ( -14.30, z-score = -0.19, R) >droWil1.scaffold_180708 5855516 104 - 12563649 GUUAUCACUUUCUGGAAUUGUGCUUUUUG-ACUGAACAGAAAUUCUUUAAAACUUUGCACGAAAUUAAAGUAAAUUGUUGCCAAAUAGUUUAGUUAAGAUUAAAC ..........(((....((((((.(((((-(..(((......))).))))))....)))))).....((.(((((((((....))))))))).)).)))...... ( -15.30, z-score = 0.19, R) >consensus GCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAAUUUAC__AGAGAUCUAAUCGGGUAUAAAAACUAAAAAUAAAU .......(((((((((.....((((......)))).......)))))))))...................................................... ( -7.18 = -8.81 + 1.64)

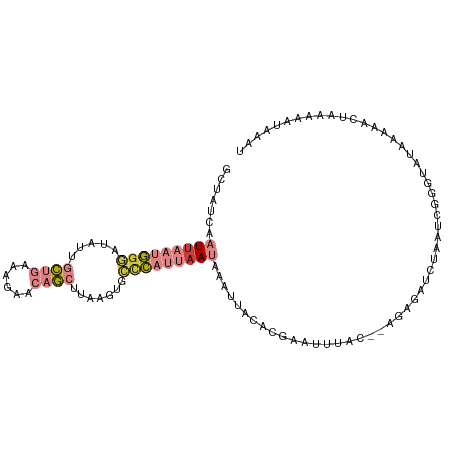

| Location | 12,836,005 – 12,836,096 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.08 |
| Shannon entropy | 0.57643 |
| G+C content | 0.34649 |
| Mean single sequence MFE | -18.15 |
| Consensus MFE | -7.56 |
| Energy contribution | -9.79 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12836005 91 + 23011544 AAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGCAAUCUAA-------UCGUGAACGCUUCGCAAAG------ ......(((.(((.(((((((((.......((((......)))).....))))))))).))).)))......-------..((((.....))))....------ ( -21.10, z-score = -2.86, R) >droEre2.scaffold_4929 14037571 91 - 26641161 UAUGUGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGCAAUCUAA-------UCAGGAACGCUUCCCGAAG------ ......(((.(((.(((((((((.......((((......)))).....))))))))).))).)))......-------((.(((.....))).))..------ ( -21.10, z-score = -2.20, R) >droYak2.chr2L 9258632 91 + 22324452 AAUUUGCGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCGAUAUCCCAUUAAUUGAUAGCAAUCUAA-------UCGUGAACGCUUCCCAAAG------ ...((((...(((.(((((((((.......((((......)))).....))))))))).))).)))).....-------.((....))..........------ ( -20.90, z-score = -2.62, R) >droSec1.super_16 999539 91 + 1878335 AAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGCAAUCUAA-------UCGUGAACGCUUCGCAAAG------ ......(((.(((.(((((((((.......((((......)))).....))))))))).))).)))......-------..((((.....))))....------ ( -21.10, z-score = -2.86, R) >droSim1.chr2L 12630837 91 + 22036055 AAAUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGCAAUCUAA-------UCGUGAACGCUUCGCAAAG------ ......(((.(((.(((((((((.......((((......)))).....))))))))).))).)))......-------..((((.....))))....------ ( -21.10, z-score = -2.86, R) >dp4.chr4_group4 485463 88 + 6586962 AAUUUAUAUAAC-UAUUUUUAGCCAUCAAAUAUUUACUUUUUCCACCAUUUUGGUUA--GCCAGCAGCCUCU-------CCGGUGCAGCCACAUGAAA------ ..(((((.....-......((((((..........................))))))--((..(((.((...-------..))))).))...))))).------ ( -9.97, z-score = 0.29, R) >droWil1.scaffold_180708 5855556 103 + 12563649 AUUUCGUGCAAAGUUUUAAAGAAUUUCUGUUCAGU-CAAAAAGCACAAUUCCAGAAAGUGAUAACAUUCUAAAUUUGAAUCAUAAACGUUCAACUAAUAGUUGU ..(((((((....(((.(..((((....))))..)-.)))..))))......((((.((....)).))))......)))...........((((.....)))). ( -11.80, z-score = 0.40, R) >consensus AAUUCGUGUAAUUUAUUAAUGGGCACUUAAGCUGUUCUUUCAGCAAUAUCCCAUUAAUUGAUAGCAAUCUAA_______UCGUGAACGCUUCGCAAAG______ ......(((.(((.(((((((((.......((((......)))).....))))))))).))).)))...................................... ( -7.56 = -9.79 + 2.23)



| Location | 12,836,005 – 12,836,096 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.08 |
| Shannon entropy | 0.57643 |
| G+C content | 0.34649 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -8.99 |
| Energy contribution | -10.00 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12836005 91 - 23011544 ------CUUUGCGAAGCGUUCACGA-------UUAGAUUGCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUU ------......((((((.((....-------...)).)))).)).(((((((((.....((((......)))).......))))))))).............. ( -20.60, z-score = -2.08, R) >droEre2.scaffold_4929 14037571 91 + 26641161 ------CUUCGGGAAGCGUUCCUGA-------UUAGAUUGCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACACAUA ------..((((((.....))))))-------...(((....))).(((((((((.....((((......)))).......))))))))).............. ( -23.10, z-score = -2.60, R) >droYak2.chr2L 9258632 91 - 22324452 ------CUUUGGGAAGCGUUCACGA-------UUAGAUUGCUAUCAAUUAAUGGGAUAUCGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACGCAAAUU ------.((((.(.((((.((....-------...)).))))....(((((((((.....((((......)))).......)))))))))......).)))).. ( -21.70, z-score = -2.05, R) >droSec1.super_16 999539 91 - 1878335 ------CUUUGCGAAGCGUUCACGA-------UUAGAUUGCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUU ------......((((((.((....-------...)).)))).)).(((((((((.....((((......)))).......))))))))).............. ( -20.60, z-score = -2.08, R) >droSim1.chr2L 12630837 91 - 22036055 ------CUUUGCGAAGCGUUCACGA-------UUAGAUUGCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAUUU ------......((((((.((....-------...)).)))).)).(((((((((.....((((......)))).......))))))))).............. ( -20.60, z-score = -2.08, R) >dp4.chr4_group4 485463 88 - 6586962 ------UUUCAUGUGGCUGCACCGG-------AGAGGCUGCUGGC--UAACCAAAAUGGUGGAAAAAGUAAAUAUUUGAUGGCUAAAAAUA-GUUAUAUAAAUU ------((((...((((((((((..-------...)).))).)))--))(((.....))).))))........((((((((((((....))-))))).))))). ( -18.70, z-score = -0.82, R) >droWil1.scaffold_180708 5855556 103 - 12563649 ACAACUAUUAGUUGAACGUUUAUGAUUCAAAUUUAGAAUGUUAUCACUUUCUGGAAUUGUGCUUUUUG-ACUGAACAGAAAUUCUUUAAAACUUUGCACGAAAU .((((.....)))).................(((((((.((....)).))))))).((((((.(((((-(..(((......))).))))))....))))))... ( -15.90, z-score = -0.47, R) >consensus ______CUUUGCGAAGCGUUCACGA_______UUAGAUUGCUAUCAAUUAAUGGGAUAUUGCUGAAAGAACAGCUUAAGUGCCCAUUAAUAAAUUACACGAAUU ............((((((.((..............)).)))).)).(((((((((.....((((......)))).......))))))))).............. ( -8.99 = -10.00 + 1.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:57 2011