
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,825,535 – 12,825,643 |
| Length | 108 |
| Max. P | 0.861126 |

| Location | 12,825,535 – 12,825,643 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Shannon entropy | 0.41787 |
| G+C content | 0.47233 |
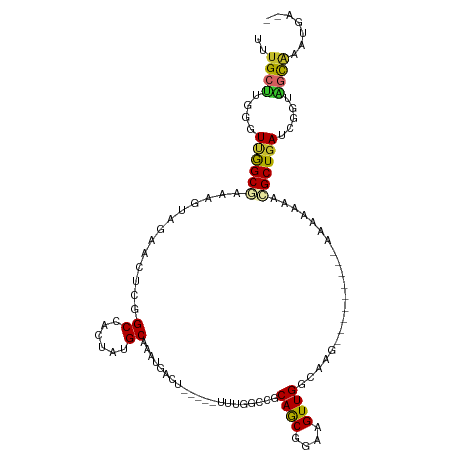
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.54 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861126 |
| Prediction | RNA |
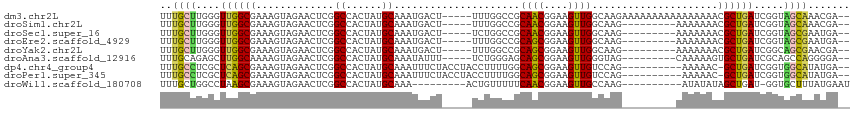
Download alignment: ClustalW | MAF
>dm3.chr2L 12825535 108 + 23011544 UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU-----UUUGGCCGCAACGGAAGUUGGCAAGAAAAAAAAAAAAAAAACGCUGAUCGGUAGCAAACGA-- (((((((.(((..(((...........((((((.....(....)...-----..))))))((((....)))).....................)))..))).).))))))...-- ( -32.50, z-score = -2.53, R) >droSim1.chr2L 12617917 99 + 22036055 UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU-----UUUGGCCGCAACGGAAGUUGGCAAG---------AAAAAAACGCUGAUCGGUAGCAAACGA-- (((((((.(((..(((...........((((((.....(....)...-----..))))))((((....)))).....---------.......)))..))).).))))))...-- ( -32.50, z-score = -2.39, R) >droSec1.super_16 989277 99 + 1878335 UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU-----UCUGGCCGCAACGGAAGUUGGCAAG---------AAAAAAACGCUGAUCGGUAGCGAAUGA-- (((((((.(((..(((...........((((((.....(....)...-----..))))))((((....)))).....---------.......)))..))).).))))))...-- ( -32.30, z-score = -2.23, R) >droEre2.scaffold_4929 14027229 99 - 26641161 UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU-----UUUGGCCGCAGCGGAAGUUGGCAAG---------AAAAAAACGCUGAUCGGUAGCGAAUGA-- (((((((.(((..(((...........((((((.....(....)...-----..))))))((((....)))).....---------.......)))..))).).))))))...-- ( -32.30, z-score = -1.90, R) >droYak2.chr2L 9247703 99 + 22324452 UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU-----UUUGGCCGCAGCGGAAGUUGGCAAG---------AAAAAAACGCUGAUCGGCAGCGAACGA-- (((((..(((((.((....))..)))))(((((.....(....)...-----..))))).((((....)))))))))---------.......(((((.....))))).....-- ( -33.50, z-score = -1.76, R) >droAna3.scaffold_12916 3580983 99 - 16180835 UUUGCAGAGCUUGGCAAAAGUAGAACUCGGCCACUAUGCAAAUAUUU-----UCUGGGAGCAGCGGAAGUUGGGUAG---------CAAAAAGUGCUGAUCGCAGCCAGGGGA-- ((((((.((..((((...((.....))..)))))).))))))..((.-----.((((..(((((....)))(..(((---------((.....)))))..)))..))))..))-- ( -32.10, z-score = -1.28, R) >dp4.chr4_group4 470947 102 + 6586962 UUUGCCUCGCUCAGCGAAAGUAGAACUCGGCCACUAUGCAAAUUUCUACCUACCUUUUGGCAGCGGAAGUUGUCCAG----------AAAAAC-GCUGAUCGGUGGCAUAUGA-- ..(((((((.((((((...((((((....((......))....)))))).....((((((((((....)))).))))----------))...)-))))).))).)))).....-- ( -39.40, z-score = -4.27, R) >droPer1.super_345 17029 102 - 19458 UUUGCCUCGCUCAGCGAAAGUAGAACUCGGCCACUAUGCAAAUUUCUACCUACCUUUUGGCAGCGGAAGUUGUCCAG----------AAAAAC-GCUGAUCGGUGGCAUAUGA-- ..(((((((.((((((...((((((....((......))....)))))).....((((((((((....)))).))))----------))...)-))))).))).)))).....-- ( -39.40, z-score = -4.27, R) >droWil1.scaffold_180708 5842842 95 + 12563649 UUUGCUGGCCUAAGCGAAAGUAGAACUCGGCCACUAUGCAAA---------ACUGUUUUUCAACGGAAGUUGCCAAG----------AUAUAUAGCUGAU-GGUGCUUUAUGAAU ((((((((((...((....)).(....))))))....)))))---------.........((((....)))).....----------.((((.(((....-...))).))))... ( -22.90, z-score = -0.71, R) >consensus UUUGCUUGGGUUGGCGAAAGUAGAACUCGGCCACUAUGCAAAUGACU_____UUUGGCCGCAGCGGAAGUUGGCAAG_________AAAAAAACGCUGAUCGGUAGCAAAUGA__ ..((((..(.((((((.............((......)).....................((((....)))).....................)))))).)...))))....... (-14.90 = -14.54 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:54 2011