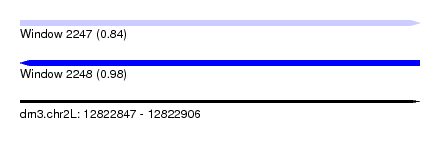
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,822,847 – 12,822,906 |
| Length | 59 |
| Max. P | 0.980724 |

| Location | 12,822,847 – 12,822,906 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 63.37 |
| Shannon entropy | 0.66138 |
| G+C content | 0.61804 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -9.31 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836269 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 12822847 59 + 23011544 -------UUUGCCACUCGGCCGCCGCCGAAGUAACUUCGGUUGCUCUUCUGCCGCUCGGCUCGCGA -------...(((....)))(((.(((((((((((....))))))..........)))))..))). ( -22.20, z-score = -1.53, R) >droSec1.super_16 986582 59 + 1878335 -------AUUGCCACUCGGCCGCCGCCGAAGCCACUUCGAUUGCUCUUCUGCCGCUCGGCUCGCGA -------...(((....)))(((.(((((.(((.....)...((......)).)))))))..))). ( -16.50, z-score = 0.20, R) >droEre2.scaffold_4929 14024594 59 - 26641161 -------UUUGCCACUCGGGCGCAGCCGAAGUCGCUUCGGCUGCUCUUCUGCCGUUCGGCUCGCGA -------...(((.....)))((((((((((...))))))))))......(((....)))...... ( -26.40, z-score = -1.86, R) >droYak2.chr2L 9244980 59 + 22324452 -------UUUGCCACUCGGCCGCAGCCGAAGCCACUUCGGCUGCUCUUCUGCCGCUCGGCUCGCGA -------...(((...((((.((((((((((...))))))))))......))))...)))...... ( -26.70, z-score = -2.52, R) >droAna3.scaffold_12916 3578250 51 - 16180835 -------------ACUCGGCUCAACCCGAAGACGCUUCGGCUACUCAUC-GUAACGCG-CUCAUAU -------------....(((.(...((((((...)))))).(((.....-)))..).)-))..... ( -8.60, z-score = 0.11, R) >droWil1.scaffold_180708 5837658 66 + 12563649 CAGAGUAAAAGCCGAACACCGAGACACGAAGCAUGGCUGGUUGCUUCGUUGCGUUGUUGUUCGUGA ............((((((.(((..(((((((((........))))))).))..))).))))))... ( -20.60, z-score = -1.05, R) >consensus _______UUUGCCACUCGGCCGCAGCCGAAGCCACUUCGGCUGCUCUUCUGCCGCUCGGCUCGCGA .....................((((((((((...))))))))))......(((....)))...... ( -9.31 = -11.20 + 1.89)

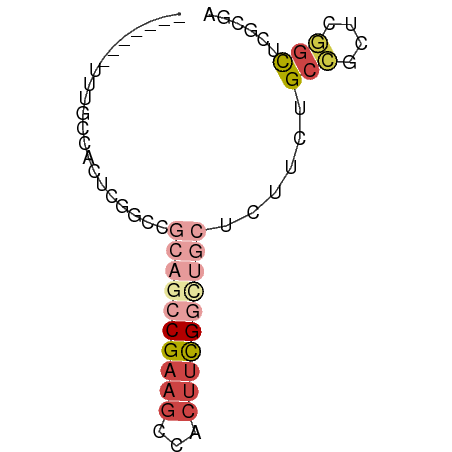
| Location | 12,822,847 – 12,822,906 |
|---|---|
| Length | 59 |
| Sequences | 6 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 63.37 |
| Shannon entropy | 0.66138 |
| G+C content | 0.61804 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -9.64 |
| Energy contribution | -12.55 |
| Covariance contribution | 2.91 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980724 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
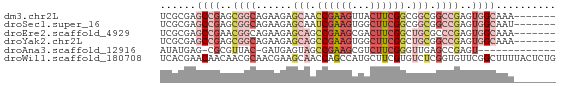
>dm3.chr2L 12822847 59 - 23011544 UCGCGAGCCGAGCGGCAGAAGAGCAACCGAAGUUACUUCGGCGGCGGCCGAGUGGCAAA------- ......((((..((((......((..((((((...))))))..)).))))..))))...------- ( -26.60, z-score = -2.21, R) >droSec1.super_16 986582 59 - 1878335 UCGCGAGCCGAGCGGCAGAAGAGCAAUCGAAGUGGCUUCGGCGGCGGCCGAGUGGCAAU------- ......((((..((((....((....))...((.((....)).)).))))..))))...------- ( -26.00, z-score = -1.77, R) >droEre2.scaffold_4929 14024594 59 + 26641161 UCGCGAGCCGAACGGCAGAAGAGCAGCCGAAGCGACUUCGGCUGCGCCCGAGUGGCAAA------- ......((((..(((.......((((((((((...))))))))))..)))..))))...------- ( -27.40, z-score = -2.27, R) >droYak2.chr2L 9244980 59 - 22324452 UCGCGAGCCGAGCGGCAGAAGAGCAGCCGAAGUGGCUUCGGCUGCGGCCGAGUGGCAAA------- ......((((..((((......((((((((((...)))))))))).))))..))))...------- ( -33.50, z-score = -3.47, R) >droAna3.scaffold_12916 3578250 51 + 16180835 AUAUGAG-CGCGUUAC-GAUGAGUAGCCGAAGCGUCUUCGGGUUGAGCCGAGU------------- ......(-((((((((-.....)))))....)))).(((((......))))).------------- ( -12.70, z-score = 0.20, R) >droWil1.scaffold_180708 5837658 66 - 12563649 UCACGAACAACAACGCAACGAAGCAACCAGCCAUGCUUCGUGUCUCGGUGUUCGGCUUUUACUCUG ...((((((....((..((((((((........))))))))....)).))))))............ ( -20.00, z-score = -2.69, R) >consensus UCGCGAGCCGAGCGGCAGAAGAGCAACCGAAGUGGCUUCGGCUGCGGCCGAGUGGCAAA_______ ......((((..((((......(((.((((((...)))))).))).))))..)))).......... ( -9.64 = -12.55 + 2.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:54 2011