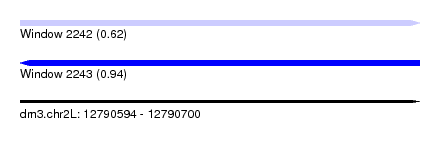
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,790,594 – 12,790,700 |
| Length | 106 |
| Max. P | 0.937648 |

| Location | 12,790,594 – 12,790,700 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.31858 |
| G+C content | 0.33859 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -11.67 |
| Energy contribution | -12.35 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12790594 106 + 23011544 --UUGUAUAAGUGUUUUUGUGCAG------UCCCCUACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG --(((((...((((..((((((..------(((......(((((........)))))..)))))))))........))))(((.(((((.....))))).))).....))))). ( -21.60, z-score = -1.50, R) >droAna3.scaffold_12916 3550824 107 - 16180835 AUGUGUUUGUGUAUUUUUGUGCAG------UCCCCAGCAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUGCCUG- ..((((......(((.((((((..------...(((...(((((........))))).))).)))))).)))....))))(((.(((((.....))))).)))..........- ( -23.20, z-score = -1.27, R) >droEre2.scaffold_4929 13994469 106 - 26641161 --AUGUGUAAGUGUUUUUGUGCAG------UCCCCUACACAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG --..(((((((.(((.((((((..------(((.......((((........))))...))))))))).))).)))))))(((.(((((.....))))).)))........... ( -25.60, z-score = -2.34, R) >droYak2.chr2L 9214855 106 + 22324452 --ACGUGUAAGUGUUUUUGUGCAG------UCCCCUAUAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG --.((((((((.(((.((((((..------(((......(((((........)))))..))))))))).))).))))))))..................(((((....))))). ( -27.80, z-score = -3.40, R) >droSec1.super_16 957818 106 + 1878335 --UUGUGUAAGUGUUUUUGUGCAG------UCCCCUACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG --..(((((((.(((.((((((..------(((......(((((........)))))..))))))))).))).)))))))(((.(((((.....))))).)))........... ( -27.10, z-score = -2.95, R) >droSim1.chr2L 12583437 106 + 22036055 --UUGUGUAAGUGUUUUUGUGCAG------UCCCCUACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG --..(((((((.(((.((((((..------(((......(((((........)))))..))))))))).))).)))))))(((.(((((.....))))).)))........... ( -27.10, z-score = -2.95, R) >droWil1.scaffold_180708 5799354 82 + 12563649 -------------------------------CCACACCAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUCCCA- -------------------------------.....((((((((........)))).))))...................(((.(((((.....))))).)))..........- ( -13.40, z-score = -3.42, R) >dp4.chr4_group4 430346 105 + 6586962 -----UGUCAUUGUGUUUGUGCAGGCCCCUCCCCCCACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUGCAAAUAUUUGCAUUUUUU---- -----.......((((((((((.((......)).(((..(((((........))))).))).))))))........))))(((.(((((.....))))).))).......---- ( -22.40, z-score = -1.44, R) >droPer1.super_68 331192 105 - 338515 -----UGUCAUUGUGUUUGUGCAGGCCCCUCCCCCCACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUGCAAAUAUUUGCAUUUUUU---- -----.......((((((((((.((......)).(((..(((((........))))).))).))))))........))))(((.(((((.....))))).))).......---- ( -22.40, z-score = -1.44, R) >consensus ___UGUGUAAGUGUUUUUGUGCAG______UCCCCUACAUAUCCAGUUAAAAGGAUAAUGGAGCACAACAAUUUUUACACGCAUAUAUUUUACAAAUAUUUGCAUUUUUGCAGG ................((((((...............(((((((........)))).)))..))))))............(((.(((((.....))))).)))........... (-11.67 = -12.35 + 0.68)

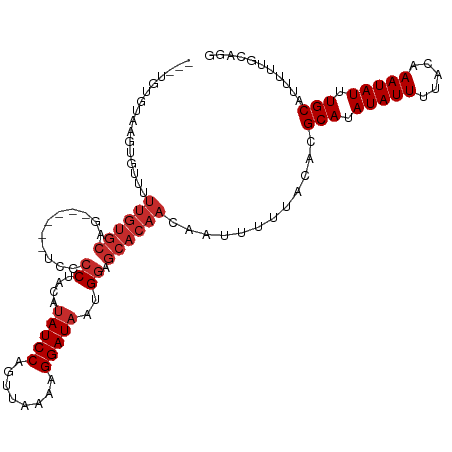
| Location | 12,790,594 – 12,790,700 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Shannon entropy | 0.31858 |
| G+C content | 0.33859 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12790594 106 - 23011544 CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUAGGGGA------CUGCACAAAAACACUUAUACAA-- ...........(((.(((((.....))))).)))((((((......(((((((((..(((((........)))))......)))------..))))))......))))))..-- ( -24.80, z-score = -2.36, R) >droAna3.scaffold_12916 3550824 107 + 16180835 -CAGGCAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGCUGGGGA------CUGCACAAAAAUACACAAACACAU -..........(((.(((((.....))))).)))(((((.....(((.((.(((((.(((((........)))))...))))))------).)))......)))))........ ( -24.30, z-score = -1.65, R) >droEre2.scaffold_4929 13994469 106 + 26641161 CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUGUGUAGGGGA------CUGCACAAAAACACUUACACAU-- ...........(((.(((((.....))))).)))((((((....((((....(((((((......))).))))(((((((....------)))))))..)))).))))))..-- ( -27.30, z-score = -2.74, R) >droYak2.chr2L 9214855 106 - 22324452 CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUAUAGGGGA------CUGCACAAAAACACUUACACGU-- ..((((((.((....)).))))))........((((((((......(((((((((..(((((........)))))......)))------..))))))......))))))))-- ( -27.70, z-score = -3.47, R) >droSec1.super_16 957818 106 - 1878335 CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUAGGGGA------CUGCACAAAAACACUUACACAA-- ...........(((.(((((.....))))).)))((((((......(((((((((..(((((........)))))......)))------..))))))......))))))..-- ( -26.70, z-score = -2.91, R) >droSim1.chr2L 12583437 106 - 22036055 CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUAGGGGA------CUGCACAAAAACACUUACACAA-- ...........(((.(((((.....))))).)))((((((......(((((((((..(((((........)))))......)))------..))))))......))))))..-- ( -26.70, z-score = -2.91, R) >droWil1.scaffold_180708 5799354 82 - 12563649 -UGGGAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGGUGUGG------------------------------- -........(((((.(((((.....))))).)))))..............((.((((.((((........)))))))).))..------------------------------- ( -16.60, z-score = -1.64, R) >dp4.chr4_group4 430346 105 - 6586962 ----AAAAAAUGCAAAUAUUUGCAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUGGGGGGAGGGGCCUGCACAAACACAAUGACA----- ----.....(((((.(((((.....))))).))))).......(..((((((((((((((((........))))).))))))(.((....)).).....)))))..)..----- ( -27.90, z-score = -1.65, R) >droPer1.super_68 331192 105 + 338515 ----AAAAAAUGCAAAUAUUUGCAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUGGGGGGAGGGGCCUGCACAAACACAAUGACA----- ----.....(((((.(((((.....))))).))))).......(..((((((((((((((((........))))).))))))(.((....)).).....)))))..)..----- ( -27.90, z-score = -1.65, R) >consensus CCUGCAAAAAUGCAAAUAUUUGUAAAAUAUAUGCGUGUAAAAAUUGUUGUGCUCCAUUAUCCUUUUAACUGGAUAUGUAGGGGA______CUGCACAAAAACACUUACACA___ .........(((((.(((((.....))))).)))))..........(((((((((..(((((........)))))......)))........))))))................ (-16.69 = -17.22 + 0.53)
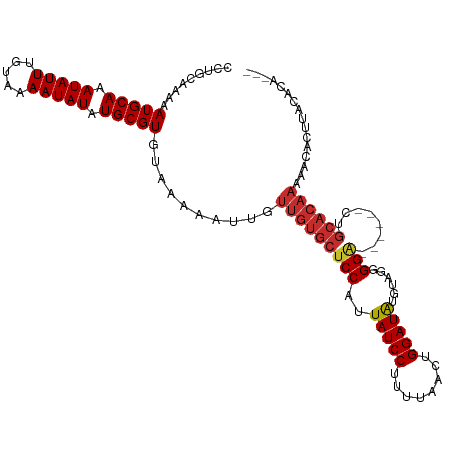
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:49 2011