| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,752,670 – 12,752,739 |
| Length | 69 |
| Max. P | 0.987199 |

| Location | 12,752,670 – 12,752,739 |
|---|---|
| Length | 69 |
| Sequences | 8 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.36882 |
| G+C content | 0.50074 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
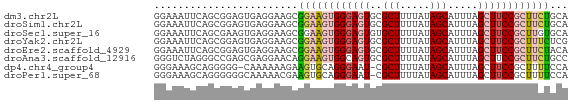
>dm3.chr2L 12752670 69 + 23011544 GGAAAUUCAGCGGAGUGAGGAAGCGGAAGUGGGAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUCUGCA ....((((....))))(.((((((((((((..((((((.........)))))).)))))))))))).). ( -28.50, z-score = -3.26, R) >droSim1.chr2L 12551176 69 + 22036055 GGAAAUUCAGCGGAGUGAGGAAGCGGAAGUGGGAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUCUGCA ....((((....))))(.((((((((((((..((((((.........)))))).)))))))))))).). ( -28.50, z-score = -3.26, R) >droSec1.super_16 927417 69 + 1878335 GGAAAUUCAGCGAAGUGAGGAAGCGGAAGUGGGAGUGUGCUUUUAUAGCAUUUAGCUUCCGCUUGUGCA .....((((......))))...(((.(((((((((((((((.....)))))...)))))))))).))). ( -25.20, z-score = -3.03, R) >droYak2.chr2L 9182398 69 + 22324452 GGAAAUUCAGCGGAGUGAGGAAGCGGAAGUGGGAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUUCUCG ....((((....))))((((((((((((((..((((((.........)))))).)))))))))))))). ( -32.40, z-score = -4.81, R) >droEre2.scaffold_4929 13963464 69 - 26641161 GGAAAUUCAGCGGAGUGAGGAAGCGGAAGUGGGAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUCUACA ....((((....))))(.((((((((((((..((((((.........)))))).)))))))))))).). ( -28.50, z-score = -3.88, R) >droAna3.scaffold_12916 3520641 69 - 16180835 GGGUCUAGGGCCGAGCGAGGAACAGGAAGUGGCAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUCUGCC .(((.....)))..((((((....((((((...(((((.........)))))..)))))).)))).)). ( -22.50, z-score = -0.14, R) >dp4.chr4_group4 376976 67 + 6586962 GGGAAAGCAGGGGG-CAAAAAAGAAGUGCAGGGAAU-CGCUUUUAUAGCAUUUAGCUUCCGCUUUUCCA ((((((((.(((((-(.......((((((...(((.-....)))...)))))).)))))))))))))). ( -24.10, z-score = -3.03, R) >droPer1.super_68 276793 68 - 338515 GGGAAAGCAGGGGGGCAAAAACGAAGUGCAGGGAAU-CGCUUUUAUAGCAUUUAGCUUCCGCUUUUCCA ((((((((..((((((.......((((((...(((.-....)))...)))))).)))))))))))))). ( -22.70, z-score = -2.35, R) >consensus GGAAAUUCAGCGGAGUGAGGAAGCGGAAGUGGGAGUGCGCUUUUAUAGCAUUUAGCUUCCGCUUCUGCA ........................((((((((((((..(((.....))).....))))))))))))... (-14.32 = -14.90 + 0.58)
| Location | 12,752,670 – 12,752,739 |
|---|---|
| Length | 69 |
| Sequences | 8 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.36882 |
| G+C content | 0.50074 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -8.76 |
| Energy contribution | -8.40 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
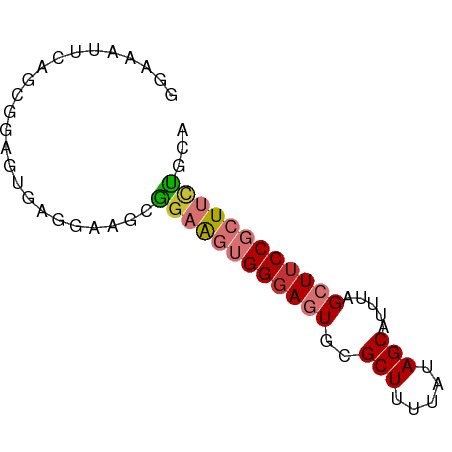
>dm3.chr2L 12752670 69 - 23011544 UGCAGAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUCCCACUUCCGCUUCCUCACUCCGCUGAAUUUCC ....((((((((((.....((((.....))))........))))))))))................... ( -17.72, z-score = -2.51, R) >droSim1.chr2L 12551176 69 - 22036055 UGCAGAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUCCCACUUCCGCUUCCUCACUCCGCUGAAUUUCC ....((((((((((.....((((.....))))........))))))))))................... ( -17.72, z-score = -2.51, R) >droSec1.super_16 927417 69 - 1878335 UGCACAAGCGGAAGCUAAAUGCUAUAAAAGCACACUCCCACUUCCGCUUCCUCACUUCGCUGAAUUUCC .....(((((((((.....((((.....))))........))))))))).................... ( -14.92, z-score = -2.76, R) >droYak2.chr2L 9182398 69 - 22324452 CGAGAAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUCCCACUUCCGCUUCCUCACUCCGCUGAAUUUCC .(((.(((((((((.....((((.....))))........))))))))).)))................ ( -19.72, z-score = -3.41, R) >droEre2.scaffold_4929 13963464 69 + 26641161 UGUAGAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUCCCACUUCCGCUUCCUCACUCCGCUGAAUUUCC ((..((((((((((.....((((.....))))........))))))))))..))............... ( -19.02, z-score = -3.30, R) >droAna3.scaffold_12916 3520641 69 + 16180835 GGCAGAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUGCCACUUCCUGUUCCUCGCUCGGCCCUAGACCC (((((.(((....)))....(((.....)))...)))))...((..(..((......))..)..))... ( -18.90, z-score = -0.76, R) >dp4.chr4_group4 376976 67 - 6586962 UGGAAAAGCGGAAGCUAAAUGCUAUAAAAGCG-AUUCCCUGCACUUCUUUUUUG-CCCCCUGCUUUCCC .(((((.((((.(((.....)))......(((-......)))............-....))))))))). ( -14.50, z-score = -1.26, R) >droPer1.super_68 276793 68 + 338515 UGGAAAAGCGGAAGCUAAAUGCUAUAAAAGCG-AUUCCCUGCACUUCGUUUUUGCCCCCCUGCUUUCCC .(((((.((((.(((.....))).((((((((-(...........))))))))).....))))))))). ( -19.10, z-score = -3.02, R) >consensus UGCAGAAGCGGAAGCUAAAUGCUAUAAAAGCGCACUCCCACUUCCGCUUCCUCACUCCGCUGAAUUUCC .((...(((....)))...((((.....))))..........................))......... ( -8.76 = -8.40 + -0.36)

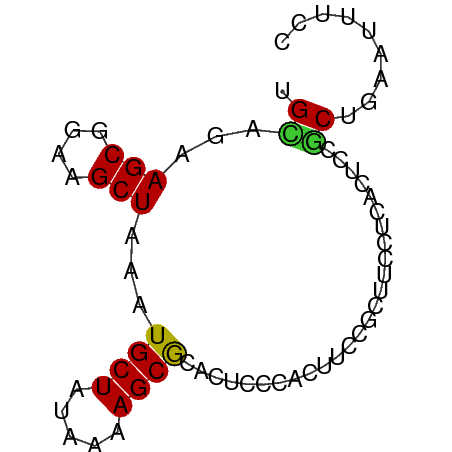
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:46 2011