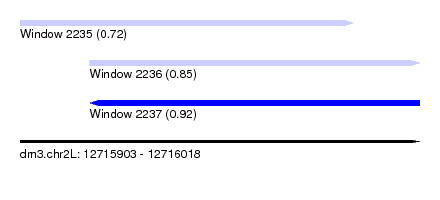
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,715,903 – 12,716,018 |
| Length | 115 |
| Max. P | 0.924315 |

| Location | 12,715,903 – 12,715,999 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.97 |
| Shannon entropy | 0.14632 |
| G+C content | 0.50266 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -29.20 |
| Energy contribution | -28.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12715903 96 + 23011544 UUUCGCCAUGGCCGCGUUGAUCUUGGUUGCCUGUAGUUUCGGUUUGAUUACCGACUGACCAGGUUUGGAUGCACCAGGAUUGGUGUUCAUAGGUUU ....(((((((..(((..(((((((((.(((((..((((((((......)))))..))))))))........)))))))))..))))))).))).. ( -31.60, z-score = -1.67, R) >droEre2.scaffold_4929 13934892 94 - 26641161 UUUCGCCAUGGCCGCGUUGUUCUUAGUUGCCUGUGGUUUCGGUUUGAUUACCGGCUGGCCAAGUUUGGAUGCACCAGGAUUGGUGAUCAUAGGU-- ....((....))................(((((((((((((((......)))))...(((((.(((((.....))))).)))))))))))))))-- ( -28.00, z-score = -0.05, R) >droYak2.chr2L 9154748 96 + 22324452 UUUCGCCAUGUCCGCGUUGUUCUUGGUUGCCUGUGGUUUAGGUUUGAUCACCGGCUGACCAGGUUUGGAUGCACCAGGAUUGGUGUUCACAGGUUU ...(((((.(((((((((...((((((((((.((((((.......)))))).))).)))))))....)))))....)))))))))........... ( -34.00, z-score = -1.81, R) >droSec1.super_16 900196 96 + 1878335 UUUUGCCGUGGCCGCGUUAAUCUUGGUUGCCUGUGGUUUUGGUUUGAUUACCGGCUGACCAGGUUUGGAUGCACCAGGAUUGGUGUUCACAGGUUU ....((((((..((((((...((((((((((.((((((.......)))))).))).)))))))....)))))(((......))))..))).))).. ( -34.20, z-score = -2.00, R) >consensus UUUCGCCAUGGCCGCGUUGAUCUUGGUUGCCUGUGGUUUCGGUUUGAUUACCGGCUGACCAGGUUUGGAUGCACCAGGAUUGGUGUUCACAGGUUU ....((((((..((((((...((((((((((.((((((.......)))))).))).)))))))....)))))(((......))))..))).))).. (-29.20 = -28.95 + -0.25)

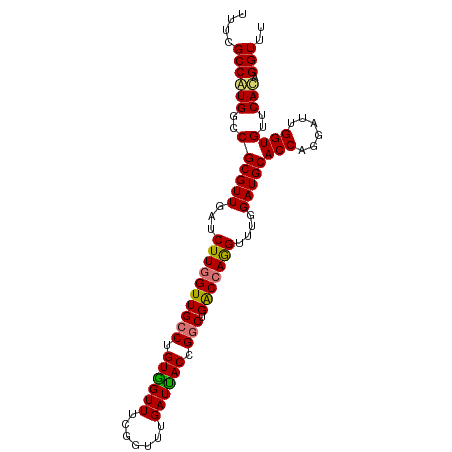
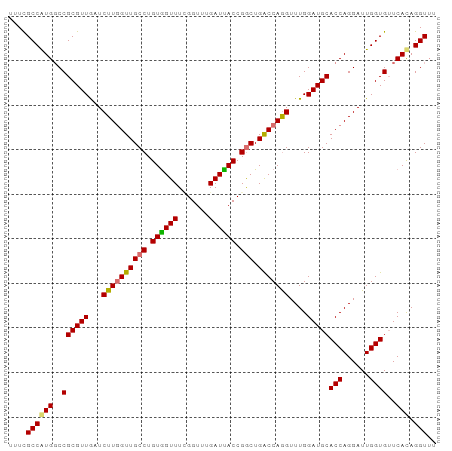
| Location | 12,715,923 – 12,716,018 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.59431 |
| G+C content | 0.46877 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -13.48 |
| Energy contribution | -16.80 |
| Covariance contribution | 3.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845526 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
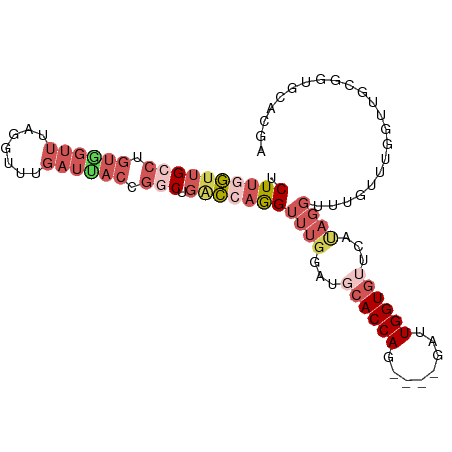
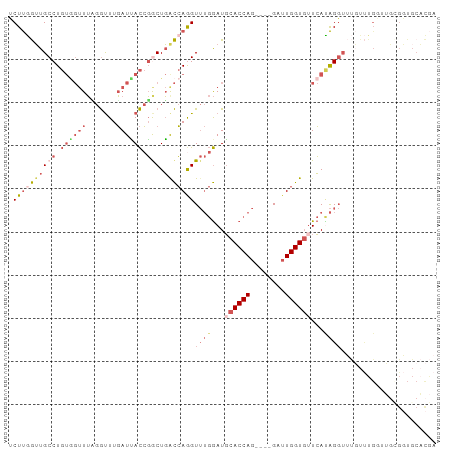
>dm3.chr2L 12715923 95 + 23011544 UCUUGGUUGCCUGUAGUUUCGGUUUGAUUACCGACUGACCAGGUUUGGAUGCACCAG----GAUUGGUGUUCAUAGGUUUGUUUGGUUGCGGUGCACGA .......(((((((((..(((((......)))))(..(.((((..((...(((((..----....)))))...))..)))).)..))))))).)))... ( -28.10, z-score = -0.75, R) >droEre2.scaffold_4929 13934912 74 - 26641161 UCUUAGUUGCCUGUGGUUUCGGUUUGAUUACCGGCUGGCCAAGUUUGGAUGCACCAG----GAUUGGUGAUCAUAGGU--------------------- ........(((((((((((((((......)))))...(((((.(((((.....))))----).)))))))))))))))--------------------- ( -26.20, z-score = -2.20, R) >droYak2.chr2L 9154768 95 + 22324452 UCUUGGUUGCCUGUGGUUUAGGUUUGAUCACCGGCUGACCAGGUUUGGAUGCACCAG----GAUUGGUGUUCACAGGUUUGUUUGCAUGAGGUGCACGA .((((((((((.((((((.......)))))).))).)))))))......((((((.(----(((....)))).((.((......)).)).))))))... ( -32.10, z-score = -1.12, R) >droSec1.super_16 900216 95 + 1878335 UCUUGGUUGCCUGUGGUUUUGGUUUGAUUACCGGCUGACCAGGUUUGGAUGCACCAG----GAUUGGUGUUCACAGGUUUGUUUGGUUGCGGUGCACGA .((((((((((.((((((.......)))))).))).)))))))......((((((.(----(((....)))).((((....)))).....))))))... ( -29.40, z-score = -0.63, R) >droGri2.scaffold_15126 4526513 91 - 8399593 GCUCAAGUGCCU-----UAAGACUUUAUAAUAUACUUAUAAAGU---AAUUUACCAGUUCCGAAUGGUUUUAAUGGCAGCUUCUGUUGCUGCUGCCUAA ((......))..-----....(((((((((.....)))))))))---.....((((........))))......((((((..(....)..))))))... ( -21.30, z-score = -1.69, R) >consensus UCUUGGUUGCCUGUGGUUUAGGUUUGAUUACCGGCUGACCAGGUUUGGAUGCACCAG____GAUUGGUGUUCAUAGGUUUGUUUGGUUGCGGUGCACGA .((((((((((.((((((.......)))))).))).)))))))((((...(((((((......)))))))...))))...................... (-13.48 = -16.80 + 3.32)
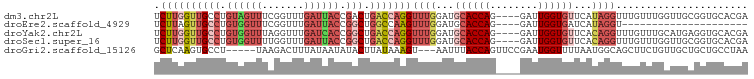
| Location | 12,715,923 – 12,716,018 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.59431 |
| G+C content | 0.46877 |
| Mean single sequence MFE | -20.63 |
| Consensus MFE | -10.04 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924315 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
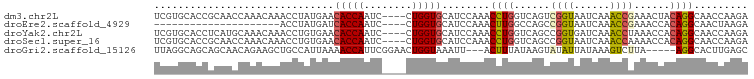
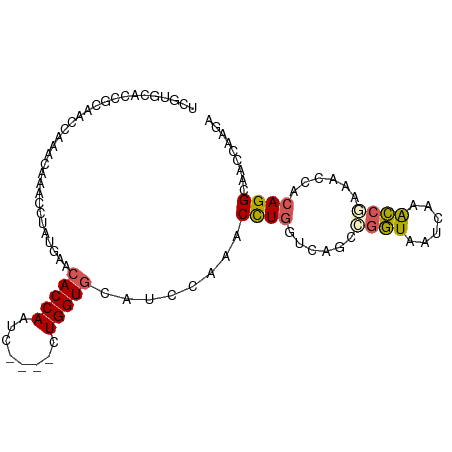
>dm3.chr2L 12715923 95 - 23011544 UCGUGCACCGCAACCAAACAAACCUAUGAACACCAAUC----CUGGUGCAUCCAAACCUGGUCAGUCGGUAAUCAAACCGAAACUACAGGCAACCAAGA ..((((((((............................----.)))))))).....((((.....(((((......))))).....))))......... ( -21.15, z-score = -1.69, R) >droEre2.scaffold_4929 13934912 74 + 26641161 ---------------------ACCUAUGAUCACCAAUC----CUGGUGCAUCCAAACUUGGCCAGCCGGUAAUCAAACCGAAACCACAGGCAACUAAGA ---------------------......((((((((...----.))))).)))....(((((...((((((......)))(....)...)))..))))). ( -17.40, z-score = -1.67, R) >droYak2.chr2L 9154768 95 - 22324452 UCGUGCACCUCAUGCAAACAAACCUGUGAACACCAAUC----CUGGUGCAUCCAAACCUGGUCAGCCGGUGAUCAAACCUAAACCACAGGCAACCAAGA ...((((.....))))......((((((..(((((...----.))))).......(((.((....)))))..............))))))......... ( -22.10, z-score = -0.97, R) >droSec1.super_16 900216 95 - 1878335 UCGUGCACCGCAACCAAACAAACCUGUGAACACCAAUC----CUGGUGCAUCCAAACCUGGUCAGCCGGUAAUCAAACCAAAACCACAGGCAACCAAGA ...(((...)))..........((((((..(((((...----.))))).......(((.((....)))))..............))))))......... ( -19.80, z-score = -0.76, R) >droGri2.scaffold_15126 4526513 91 + 8399593 UUAGGCAGCAGCAACAGAAGCUGCCAUUAAAACCAUUCGGAACUGGUAAAUU---ACUUUAUAAGUAUAUUAUAAAGUCUUA-----AGGCACUUGAGC ...((((((..(....)..))))))......((((........)))).....---(((((((((.....)))))))))((((-----((...)))))). ( -22.70, z-score = -2.38, R) >consensus UCGUGCACCGCAACCAAACAAACCUAUGAACACCAAUC____CUGGUGCAUCCAAACCUGGUCAGCCGGUAAUCAAACCGAAACCACAGGCAACCAAGA ..............................(((((........)))))........((((......((((......))))......))))......... (-10.04 = -10.16 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:44 2011