
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,702,571 – 12,702,686 |
| Length | 115 |
| Max. P | 0.619171 |

| Location | 12,702,571 – 12,702,685 |
|---|---|
| Length | 114 |
| Sequences | 15 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.39 |
| Shannon entropy | 0.76172 |
| G+C content | 0.56650 |
| Mean single sequence MFE | -42.05 |
| Consensus MFE | -13.22 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12702571 114 + 23011544 AGGAUACCCCUGGGUGGCUUGAUACCCAGGCGUUGGAACUUAUCCGCGUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUGGCCAAACGUAGCUCGGAUUGACCGCCAAUGUCG-- (((.....)))(((((......))))).((((((((.....((((((((((((((..(((((.....))))))))))))))((........)).)))))......)))))))).-- ( -49.40, z-score = -2.49, R) >droSim1.chr2L 12511098 114 + 22036055 AGGAUACCCCUGGGUGGCUUGAUACCCAGGCGUUGGAACUUUUCCGCGUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAUGUCG-- (((.....)))(((((......))))).((((((((.....((((((((((((((..(((((.....)))))))))))).))).....))))..(((.....))))))))))).-- ( -47.10, z-score = -1.88, R) >droSec1.super_16 886892 114 + 1878335 AGAAUACCCCUGGGUGGCUUAAUACCCAGGCGUUGGAACUUUUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAUGUCG-- .((.....((((((((......))))))))((((((.....((((((((((((((..(((((.....))))))))))))).)).....))))..(((.....))))))))))).-- ( -45.80, z-score = -2.47, R) >droYak2.chr2L 9141195 114 + 22324452 AGGAUGCCCCUUGGCGGCUUGAUGCCCAGACGUUGGAACUUCUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAUAUCC-- .(((((....(((((((.(..((.((.((.(((((((.....)))((((((((((..(((((.....))))))))))))).))..))))...)).))))..)))))))))))))-- ( -49.70, z-score = -3.06, R) >droEre2.scaffold_4929 13921496 114 - 26641161 AGGAUACCCCUUGGGGGCUUGAUACCCAGGCGUUGGAACUUAUCCGCAUGCAGCAGAGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGCUUGACCGCCAAUGUCU-- .((((((((....)))(((((.....))))).........)))))((((((((((..(((((.....))))))))))))).))............(((......))).......-- ( -44.20, z-score = -1.08, R) >droAna3.scaffold_12916 3478269 114 - 16180835 AGGAUUCCUCUGGGUGGCUUGAUGCCCAAUCGCUGGAACUUUUCCGCGUGGAGCAGCGGCCACUCAAUCGCCUGCUGCAUCGCCAAACGGAGCUCGGACUGACCCCCGAUAUCC-- .((((..(((((..((((..((((((((..(((.(((.....)))))))))((((((((........))).))))))))))))))..))))).((((........)))).))))-- ( -44.00, z-score = -1.73, R) >dp4.chr4_group4 334302 114 + 6586962 AGAAUGCCACGCGGUGGCUUAAUGCCAAGUCGCUGGAACUUCUCCGCAUGAAGCAGUGGCCAUUCGAUAGCCUGUUGCAUGGCCAGACGGAGUUCGGACUGGCCGCCAAUGUCU-- .(((((((((((((((((.....))))..)))).(((.....)))((.....)).)))).)))))((((((.....))..((((((.(.(....).).)))))).....)))).-- ( -42.10, z-score = -0.78, R) >droPer1.super_68 234436 114 - 338515 AGAAUGCCACGCGGUGGCUUAAUGCCAAGUCGCUGGAACUUCUCCGCAUGAAGCAGUGGCCAUUCGAUAGCCUGUUGCAUGGCCAGACGGAGCUCGGACUGGCCGCCAAUGUCC-- .(((((((((((((((((.....))))..)))).(((.....)))((.....)).)))).)))))((((((.....))..((((((.(.(....).).)))))).....)))).-- ( -41.70, z-score = -0.50, R) >droWil1.scaffold_180708 9027941 114 + 12563649 AGUAAUCCUCUAGGAGGUUUGAUGCCCAAGCGUUGAAAUUUCUCGGCAUGCAACAGAGGCCAUUCAAUGGCUUGUUGAAGAGCCAAACGCAAUUGGGAUUGUCCUCCUAUGUCG-- ..........(((((((..((((.((((((((((((......))(((...(((((..((((((...)))))))))))....))).)))))..))))))))).))))))).....-- ( -40.10, z-score = -2.88, R) >droVir3.scaffold_13246 631297 114 - 2672878 AGCACGCCGCGCGGCGGCUUGAUGCCCAGGCGCUGGAAUUUAUCCGCAUGCAGCAGCGGCCACUCGAUGGCCUGCUGCAGUGCCAGCCGCAGCUCCGCCUGGCCGCCAAUGUCC-- .((.....))((((((((.....)))((((((..(((.....)))((.(((.((((((((((.....))))).))))).((....)).)))))..)))))))))))........-- ( -52.10, z-score = -0.07, R) >droMoj3.scaffold_6500 6214871 116 + 32352404 AGCAAGCCCUUCGGCAGGCUCAAGCCAAAGCGGGCAUGGGCCUCGCUGUGGCGCAGAGCAGCCAAGACGGAGACCUGCAGUGUGCGCUUCAGUGCAGCCAUGCCGCCAAUCGACUC .((..(((....))).(((....)))...)).(((((((.(..(((((.((((((.(((((((.....))....))))..).)))))).)))))..))))))))............ ( -47.40, z-score = 0.24, R) >droGri2.scaffold_15252 15244602 114 + 17193109 AGUACGCCACGUGGUGGUUUAAUGCCCAAACGCUCGAAUUUAUCCGCAUGGAGCAGUGGCCAUUCGAUGGCCUGUUGAAGGGUCAAACGUAGUGCGGAUUGGCCACCAAUAUCC-- .((...(((.((((.(((((...((......))..)))))...)))).))).)).(((((((.((..((((((......))))))..((.....)))).)))))))........-- ( -36.30, z-score = -0.34, R) >anoGam1.chr2R 33998375 114 + 62725911 AUCACACCCUUCGGCGGUUUGAUGCCCAUCUCCUCGUAGUACUCGGGGUGCGUGAGGGGCAGCUCGACCGAUUCCUUGAUCUCCUGGAUCUGCGUGUCCAGCCCGCCGAUGUCG-- ..........(((((((.....(((((..(.(((((.......))))).)(....))))))(((.(((((..(((..(....)..)))....)).))).)))))))))).....-- ( -42.10, z-score = -0.66, R) >apiMel3.Group4 1842022 114 + 10796202 AAAACACCUUUAGGUGGAGUUAUACCCAUUCUGAAAAAUACUUCAGGAUGACAUAAUGGCCAUUCAAUAGCUUGUUUUAAUUUUAAUUUUAAGUCUUUUUGUCCUCCUAUAUCU-- ..........((((.((((((((...(((((((((......)))))))))..))))).......(((.((((((..(((....)))...)))).))..)))))).)))).....-- ( -21.00, z-score = -1.22, R) >triCas2.ChLG4 2593415 114 - 13894384 AAAACCCCCUUCGGUGGGGUCACCCCUAGUCGUAGAAAACUCUCAGGGUGUCUCAAAGGCCACUCUACAGCCUGUCUGAGUAUCAAUUUAAGAUUCUGAAGCCCCCCAAUAUCA-- ............((.((((((((((..((..((.....)).))..))))).((((.((((.........))))...))))....................))))))).......-- ( -27.70, z-score = -0.57, R) >consensus AGAAUACCCCUCGGUGGCUUGAUGCCCAGGCGUUGGAACUUCUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAUGUCC__ ............(((((...................................((((((((.........))).))))).....(((.(........).))).)))))......... (-13.22 = -12.73 + -0.49)

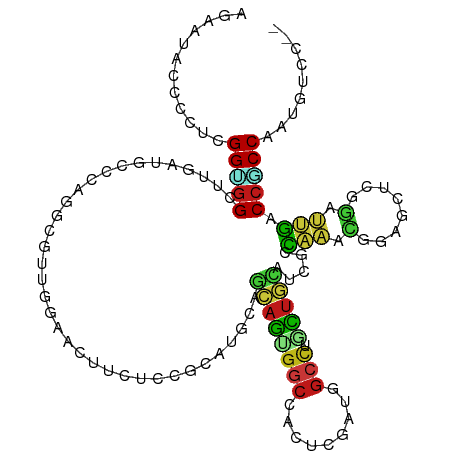
| Location | 12,702,572 – 12,702,686 |
|---|---|
| Length | 114 |
| Sequences | 15 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.31 |
| Shannon entropy | 0.76252 |
| G+C content | 0.57526 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -13.22 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12702572 114 + 23011544 GGAUACCCCUGGGUGGCUUGAUACCCAGGCGUUGGAACUUAUCCGCGUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUGGCCAAACGUAGCUCGGAUUGACCGCCAAU--GUCGG .....((((((((((......))))))))((((((.....((((((((((((((..(((((.....))))))))))))))((........)).)))))......)))))--)..)) ( -49.60, z-score = -2.31, R) >droSim1.chr2L 12511099 114 + 22036055 GGAUACCCCUGGGUGGCUUGAUACCCAGGCGUUGGAACUUUUCCGCGUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAU--GUCGG .....((((((((((......))))))))((((((.....((((((((((((((..(((((.....)))))))))))).))).....))))..(((.....))))))))--)..)) ( -47.30, z-score = -1.71, R) >droSec1.super_16 886893 114 + 1878335 GAAUACCCCUGGGUGGCUUAAUACCCAGGCGUUGGAACUUUUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAU--GUCGG .....((((((((((......))))))))((((((.....((((((((((((((..(((((.....))))))))))))).)).....))))..(((.....))))))))--)..)) ( -45.90, z-score = -2.26, R) >droYak2.chr2L 9141196 114 + 22324452 GGAUGCCCCUUGGCGGCUUGAUGCCCAGACGUUGGAACUUCUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAU--AUCCG (((((....(((((((.(..((.((.((.(((((((.....)))((((((((((..(((((.....))))))))))))).))..))))...)).))))..)))))))))--)))). ( -51.20, z-score = -3.60, R) >droEre2.scaffold_4929 13921497 114 - 26641161 GGAUACCCCUUGGGGGCUUGAUACCCAGGCGUUGGAACUUAUCCGCAUGCAGCAGAGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGCUUGACCGCCAAU--GUCUG ((((((((....)))(((((.....))))).........)))))((((((((((..(((((.....))))))))))))).))....((((....(((......)))...--.)))) ( -45.40, z-score = -1.64, R) >droAna3.scaffold_12916 3478270 114 - 16180835 GGAUUCCUCUGGGUGGCUUGAUGCCCAAUCGCUGGAACUUUUCCGCGUGGAGCAGCGGCCACUCAAUCGCCUGCUGCAUCGCCAAACGGAGCUCGGACUGACCCCCGAU--AUCCG ((((..(((((..((((..((((((((..(((.(((.....)))))))))((((((((........))).))))))))))))))..))))).((((........)))).--)))). ( -45.50, z-score = -2.35, R) >dp4.chr4_group4 334303 114 + 6586962 GAAUGCCACGCGGUGGCUUAAUGCCAAGUCGCUGGAACUUCUCCGCAUGAAGCAGUGGCCAUUCGAUAGCCUGUUGCAUGGCCAGACGGAGUUCGGACUGGCCGCCAAU--GUCUG (((((((((((((((((.....))))..)))).(((.....)))((.....)).)))).)))))((((((.....))..((((((.(.(....).).)))))).....)--))).. ( -41.90, z-score = -0.80, R) >droPer1.super_68 234437 114 - 338515 GAAUGCCACGCGGUGGCUUAAUGCCAAGUCGCUGGAACUUCUCCGCAUGAAGCAGUGGCCAUUCGAUAGCCUGUUGCAUGGCCAGACGGAGCUCGGACUGGCCGCCAAU--GUCCG (((((((((((((((((.....))))..)))).(((.....)))((.....)).)))).)))))((((((.....))..((((((.(.(....).).)))))).....)--))).. ( -41.50, z-score = -0.40, R) >droWil1.scaffold_180708 9027942 114 + 12563649 GUAAUCCUCUAGGAGGUUUGAUGCCCAAGCGUUGAAAUUUCUCGGCAUGCAACAGAGGCCAUUCAAUGGCUUGUUGAAGAGCCAAACGCAAUUGGGAUUGUCCUCCUAU--GUCGG .........(((((((..((((.((((((((((((......))(((...(((((..((((((...)))))))))))....))).)))))..))))))))).))))))).--..... ( -40.10, z-score = -2.60, R) >droVir3.scaffold_13246 631298 114 - 2672878 GCACGCCGCGCGGCGGCUUGAUGCCCAGGCGCUGGAAUUUAUCCGCAUGCAGCAGCGGCCACUCGAUGGCCUGCUGCAGUGCCAGCCGCAGCUCCGCCUGGCCGCCAAU--GUCCG ((.....))((((((((.....)))((((((..(((.....)))((.(((.((((((((((.....))))).))))).((....)).)))))..)))))))))))....--..... ( -51.90, z-score = -0.08, R) >droMoj3.scaffold_6500 6214872 116 + 32352404 GCAAGCCCUUCGGCAGGCUCAAGCCAAAGCGGGCAUGGGCCUCGCUGUGGCGCAGAGCAGCCAAGACGGAGACCUGCAGUGUGCGCUUCAGUGCAGCCAUGCCGCCAAUCGACUCG ((..(((....))).(((....)))...)).(((((((.(..(((((.((((((.(((((((.....))....))))..).)))))).)))))..))))))))............. ( -47.20, z-score = 0.34, R) >droGri2.scaffold_15252 15244603 114 + 17193109 GUACGCCACGUGGUGGUUUAAUGCCCAAACGCUCGAAUUUAUCCGCAUGGAGCAGUGGCCAUUCGAUGGCCUGUUGAAGGGUCAAACGUAGUGCGGAUUGGCCACCAAU--AUCCG ...((.....((((((((....((......))........((((((((...((..(((((.((((((.....)))))).)))))...)).)))))))).))))))))..--...)) ( -36.40, z-score = -0.22, R) >anoGam1.chr2R 33998376 114 + 62725911 UCACACCCUUCGGCGGUUUGAUGCCCAUCUCCUCGUAGUACUCGGGGUGCGUGAGGGGCAGCUCGACCGAUUCCUUGAUCUCCUGGAUCUGCGUGUCCAGCCCGCCGAU--GUCGG .....((..(((((((.....(((((..(.(((((.......))))).)(....))))))(((.(((((..(((..(....)..)))....)).))).)))))))))).--...)) ( -42.60, z-score = -0.40, R) >apiMel3.Group4 1842023 114 + 10796202 AAACACCUUUAGGUGGAGUUAUACCCAUUCUGAAAAAUACUUCAGGAUGACAUAAUGGCCAUUCAAUAGCUUGUUUUAAUUUUAAUUUUAAGUCUUUUUGUCCUCCUAU--AUCUG .........((((.((((((((...(((((((((......)))))))))..))))).......(((.((((((..(((....)))...)))).))..)))))).)))).--..... ( -21.00, z-score = -1.21, R) >triCas2.ChLG4 2593416 114 - 13894384 AAACCCCCUUCGGUGGGGUCACCCCUAGUCGUAGAAAACUCUCAGGGUGUCUCAAAGGCCACUCUACAGCCUGUCUGAGUAUCAAUUUAAGAUUCUGAAGCCCCCCAAU--AUCAC ...........((.((((((((((..((..((.....)).))..))))).((((.((((.........))))...))))....................)))))))...--..... ( -27.70, z-score = -0.52, R) >consensus GAAUACCCCUCGGUGGCUUGAUGCCCAGGCGUUGGAACUUCUCCGCAUGCAGCAGUGGCCACUCGAUGGCCUGCUGCAUCGCCAAACGGAGCUCGGAUUGACCGCCAAU__GUCCG ...........(((((...................................((((((((.........))).))))).....(((.(........).))).))))).......... (-13.22 = -12.73 + -0.49)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:41 2011