
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,192,397 – 1,192,517 |
| Length | 120 |
| Max. P | 0.906065 |

| Location | 1,192,397 – 1,192,517 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.48961 |
| G+C content | 0.57778 |
| Mean single sequence MFE | -47.64 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.906065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
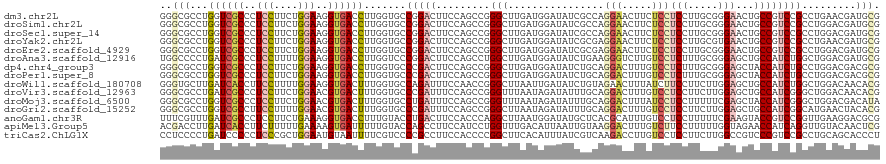
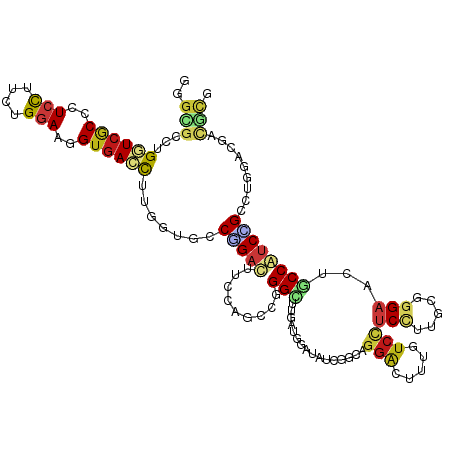
>dm3.chr2L 1192397 120 - 23011544 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGCCAGGAACUUCUCCUCCUUGCGGGAACUGCCGUCCGCCUGAACGAUGCG .((((((.(((((((.(((....))).)))))))..))))))((....))...(((((..(((((...((((.((((........)))).))))......))))).)))))......... ( -56.70, z-score = -3.43, R) >droSim1.chr2L 1171796 120 - 22036055 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGCCAGGAACUUCUCCUCCUUGCGGGAACUGCCGUCCGCCUGGACGAUGCG .((((((.(((((((.(((....))).)))))))..))))))((....)).(((((((..(((((...((((.((((........)))).))))......))))).)))))).)...... ( -60.10, z-score = -3.81, R) >droSec1.super_14 1152413 120 - 2068291 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGCCAGGAACUUCUCCUCCUUGCGGGAACUGCCGUCCGCCUGGACGAUGCG .((((((.(((((((.(((....))).)))))))..))))))((....)).(((((((..(((((...((((.((((........)))).))))......))))).)))))).)...... ( -60.10, z-score = -3.81, R) >droYak2.chr2L 1172729 120 - 22324452 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGCGAGGAACUUCUCCUCCUUGCGUGAACUGCCGUCCGCCUGAACGAUGCG .((((((.(((((((.(((....))).)))))))..))))))((....))...(((((..(((((....((((((((........)))))))).......))))).)))))......... ( -59.10, z-score = -4.86, R) >droEre2.scaffold_4929 1237856 120 - 26641161 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGCGAGGAACUUCUCCUCCUUGCGGGAACUGCCGUCCGCCUGGACGAUGCG .((((((.(((((((.(((....))).)))))))..))))))((....)).(((((((..(((((...(((((((((........)))))))))......))))).)))))).)...... ( -63.90, z-score = -4.82, R) >droAna3.scaffold_12916 4664510 120 - 16180835 UGGCCCCUGAUCGCCCUCCUUUUGGAAGGUGACCUUGGUCCCGGACUUCCAGCCUGGCUUGAUGGAUAUCUGAAGGGUCUUGUCCUCUUUGCGGGAGCUGCCAUCUGGCUGGACGAUGCG .((..((.(.(((((.(((....))).))))).)..))..))...(.(((((((((((..((.(((((...((....)).)))))))...((....)).))))...))))))).)..... ( -43.20, z-score = -0.36, R) >dp4.chr4_group3 9645934 120 - 11692001 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCCGACUUCCAGCCGGGCUUGAUGGAUAUCUGCAGGACUUUGUCCUCUUUGCGGGAGCUACCAUCUGCCUGGACGACGCG (((((((.(((((((.(((....))).)))))))..)))))))(.(.((...((((((..(((((...((((((((((...))))....)))))).....))))).))))))..)).)). ( -61.30, z-score = -4.94, R) >droPer1.super_8 844766 120 - 3966273 GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCCGACUUCCAGCCGGGCUUGAUGGAUAUCUGCAGGACUUUGUCCUCUUUGCGGGAGCUACCAUCUGCCUGGACGACGCG (((((((.(((((((.(((....))).)))))))..)))))))(.(.((...((((((..(((((...((((((((((...))))....)))))).....))))).))))))..)).)). ( -61.30, z-score = -4.94, R) >droWil1.scaffold_180708 2792371 120 - 12563649 GGGUGCUUGAUCACCUUCCUUUUGGAAGGUGACUUUGGUGCCAGAUUUCCAACCGGGCUUAAUUGAUAUCUGUAGAACUUUAUCUUCCUUCUUGGAGCUGCCAUCUGGCUGGACAACACG .((..((.(((((((((((....))))))))).)).))..)).....((((.((.(((......((((...(.....)..))))((((.....))))..)))....)).))))....... ( -38.80, z-score = -2.50, R) >droVir3.scaffold_12963 7119472 120 - 20206255 GGGCGCCUGAUCGCCCUCCUUCUGGAACGUGACUUUGGUGCCCGAUUUCCAGCCGGGUUUAAUAGAUAUUUGCAGGACUUUGUCCUCCUUCUUGGAGCUGCCAUCGGGCUGGACAACACG (((((((.((((((..(((....)))..)))).)).)))))))....(((((((.(((.............(((((((...)))((((.....)))))))).))).)))))))....... ( -46.34, z-score = -2.68, R) >droMoj3.scaffold_6500 28546959 120 + 32352404 GGGCGCCUGGUCGCCCUCCUUCUGGAACGUGACUUUGGUGCCUGAUUUCCAGCCGGGUUUAAUAGAUAUUUGCAGGACUUUAUCCUCCUUUUUCGAGCUACCAUCGGGCUGGACGACAUA (((((((.((((((..(((....)))..))))))..)))))))....(((((((.(((....(((........((((........))))........)))..))).)))))))....... ( -44.69, z-score = -2.82, R) >droGri2.scaffold_15252 10269104 120 - 17193109 GGGCGCCUGGUCGCCUUCCUUUUGGAACGUGACUUUGGUGCCCGAUUUCCAGCCGGGCUUAAUAGAUAUUUGCAGGACUUUGUCCUCCUUCUUGGAGCUGCCAUCGGCAUGAACUACACG (((((((.((((((.((((....)))).))))))..)))))))........(((((((((...(((.....(.(((((...))))).).)))..))))).....))))............ ( -47.30, z-score = -3.56, R) >anoGam1.chr3R 52095118 120 - 53272125 UUUCGUUUGAUCGCCCUCCUUCUGAAAGGUGACCUUUGUACCUGACUUCCACCCAGGCUUAAUGGAUAUGCUCACGCAUUUGUCCUCCUUUUUCGAAGUACCGUCCGGUUGAAGGACGCG ..((.((..((((..(..((((.((((((.((...(((..((((.........))))..))).((((((((....))))..)))))))))))).))))....)..))))..)).)).... ( -28.10, z-score = -0.38, R) >apiMel3.Group5 8466668 120 + 13386189 ACGACCUUGAUCACCUUCUUUUUGAAAAGUGAUUUUUGUACCAGCCUUCCAUCCUGGUUUGACAUUAAUUGUAAGGACUUUGUCUUCCUUUUUGGUAGAACCAUCAGGUUGUACAACUCG .(((....((((((.(((.....)))..)))))).((((((.(((((.......((((((...(((((....(((((........))))).))))).))))))..))))))))))).))) ( -26.30, z-score = -1.18, R) >triCas2.ChLG1X 3376656 120 - 8109244 CCUCCCCUGAUCCCCCUCCCGCUGGAAUGUAAUUUUCGUCCCCGCCUUCCACCCCGGCUUCACAUUUAUCGUCAAGACCUUGUCCUCCUUCUUGGCCGUCCCGUCCGCCUGCAGCACCCU ....................((((((((((.......(....)(((.........)))...))))))...(((((((............)))))))...............))))..... ( -17.30, z-score = -0.03, R) >consensus GGGCGCCUGGUCGCCCUCCUUCUGGAAGGUGACCUUGGUGCCGGACUUCCAGCCGGGCUUGAUGGAUAUCGGCAGGACUUUGUCCUCCUUGCGGGAACUGCCAUCCGCCUGGACGACGCG ..(((...((((((..(((....)))..)))))).......(((((.........(((................(((.....)))(((.....)))...)))))))).........))). (-14.06 = -14.87 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:50 2011