| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,674,372 – 12,674,465 |
| Length | 93 |
| Max. P | 0.983260 |

| Location | 12,674,372 – 12,674,465 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 66.09 |
| Shannon entropy | 0.66340 |
| G+C content | 0.50187 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12674372 93 + 23011544 ACCACCCGCACCAACAAUGGGCGAAAAGGGAAAAUCCAAAGUGAAAAGCUCUGA----CGC-------AAAACUGGUGUCACUACCAACUGUGACCCGGUACCC-- ......(((.(((....))))))....(((......((.(((.....))).)).----...-------...(((((.(((((........)))))))))).)))-- ( -26.50, z-score = -2.00, R) >droVir3.scaffold_13246 2002725 92 + 2672878 -----ACCCUUAGAAAAUGUAAAAAAAAAAAUAAGUAAAUCCGGAAAAAAAUAAAAG-CUC--------ACGCUGGUGUCAAUACCAACUGUGCCCCUGCAGGCUC -----....................................(((...........((-(..--------..)))((((....))))..))).(((......))).. ( -9.60, z-score = 0.85, R) >droPer1.super_1 4899624 85 + 10282868 ----AUCACCCUCACAAAA----AGUAAAGUAAAAUCCAAAGGGAAAGCUCUAA----CAC-------GGCGCUGGUGUCACUACCAACUGUGACCCGGUGCUC-- ----....((((.......----.................))))..........----...-------((((((((.(((((........))))))))))))).-- ( -23.96, z-score = -2.60, R) >dp4.chr4_group3 3423005 85 + 11692001 ----AUCACCCUCACAAAA----AGUAAAGUAAAAUCCAAAGGGAAAGCUCUAA----CAC-------GGCGCUGGUGUCACUACCAACUGUGACCCGGUGCUC-- ----....((((.......----.................))))..........----...-------((((((((.(((((........))))))))))))).-- ( -23.96, z-score = -2.60, R) >droAna3.scaffold_12916 3448448 74 - 16180835 ------------------ACCCUGGCAGAGGGAGACGCGACAGGAAAGCUCUGA----CUC--------AAACUGGUGUCACUACCAACUGUGACCCGGUACUC-- ------------------.........(((..(((.((.........)))))..----)))--------..(((((.(((((........))))))))))....-- ( -23.20, z-score = -0.74, R) >droEre2.scaffold_4929 13892583 95 - 26641161 --ACCCCGCGCCAACAAUGGGCGAGCAGGGAAAAUCCAAAGUGAAAAGCUCUGAAAACUGU-------GAAACUGGUGUCACUACCAACUGUGACCCGGUACCC-- --..(((.((((.......))))....)))......((.(((.....))).))........-------...(((((.(((((........))))))))))....-- ( -27.40, z-score = -1.57, R) >droYak2.chr2L 9112558 102 + 22324452 --ACCCCGCACCAACAAUGGGCCAAAUGGGAAAAUCCAAAGUGAAAAGCUUGGAAAACUGCUGCCCUCGAAACUGGUGUCACUACCAACUGCGACCCGGUACCC-- --.....((.(((....))))).....(((....(((((.((.....))))))).................(((((.(((.(........).)))))))).)))-- ( -23.40, z-score = 0.01, R) >droSec1.super_16 858667 93 + 1878335 ACCACCCGCACCAACAAUGGGCGAAAAGGGAAAAUCCAAAGUGAAAAGCUCUGA----CGC-------AAAACUGGUGUCACUACCAACUGUGACCCGGUACCC-- ......(((.(((....))))))....(((......((.(((.....))).)).----...-------...(((((.(((((........)))))))))).)))-- ( -26.50, z-score = -2.00, R) >droSim1.chr2L 12482813 93 + 22036055 ACCACCCGCACCAACAAUGGGCGAAAAGGGAAAAUCCAAAGUGAAAAGCUCUGA----CGC-------AAAACUGGUGUCACUACCAACUGUGACCCGGUACCC-- ......(((.(((....))))))....(((......((.(((.....))).)).----...-------...(((((.(((((........)))))))))).)))-- ( -26.50, z-score = -2.00, R) >consensus ____CCCGCACCAACAAUGGGCGAAAAGGGAAAAUCCAAAGUGAAAAGCUCUGA____CGC_______GAAACUGGUGUCACUACCAACUGUGACCCGGUACCC__ .......................................................................(((((.(((((........))))))))))...... (-12.70 = -12.82 + 0.12)

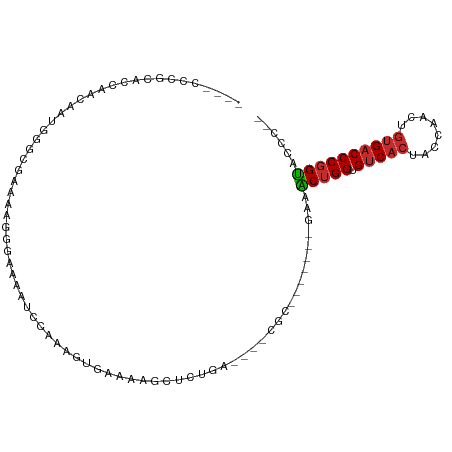
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:38 2011