
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,646,713 – 12,646,812 |
| Length | 99 |
| Max. P | 0.773554 |

| Location | 12,646,713 – 12,646,812 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.47721 |
| G+C content | 0.57962 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -22.16 |
| Energy contribution | -24.56 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773554 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 12646713 99 - 23011544 GGGCCUGUCAAAGUCUUGAGCUCCUCAGCUGUCUUGGUAUCGUCAUC---GUAUGGGGAUCGGCGGCAUCUUGCUGCCGAGCUUUGCCAGUGUCCACUGGUC---- (((((((.((((((....((((....)))).((((.(((.((....)---)))).))))((((((((.....)))))))))))))).))).)))).......---- ( -38.70, z-score = -2.02, R) >droSim1.chr2L 12454500 99 - 22036055 GGGCCUGUCAAAGUCUUGAGCUCCUCAGCUGUCUUGGUAUCGUCAUC---GUAUGGGGAUCGGCGGCAUCUUGCUGCCGAGCUUUGCCAGUGUCCACUGGUC---- (((((((.((((((....((((....)))).((((.(((.((....)---)))).))))((((((((.....)))))))))))))).))).)))).......---- ( -38.70, z-score = -2.02, R) >droSec1.super_16 831668 99 - 1878335 GGGCCUGUCAAAGUCUUGAGCUCCUCAGCUGUCUUGGUAUCGUCAUC---GUAUGGGGAUCGGCGGCAUCUUGCUGCCGAGCUUUGCCAGUGUCCACUGGUC---- (((((((.((((((....((((....)))).((((.(((.((....)---)))).))))((((((((.....)))))))))))))).))).)))).......---- ( -38.70, z-score = -2.02, R) >droYak2.chr2L 9083998 102 - 22324452 GGGCCUGUCAAAGUCUUGAGCUCUUCAGAUGUCUUGGUAUCGGGGAUCGGGCAUGGGGAUCGGCGGCAUGGUGCUGCCGAGCUUUGCCAGUGUCCACUGGUC---- (((((((.((((((((((.((((.((.((((......))))...))..)))).))))..((((((((.....)))))))))))))).))).)))).......---- ( -39.80, z-score = -0.77, R) >droEre2.scaffold_4929 13863779 102 + 26641161 GGGCCUGUCAAAGUCUGGAGCUCUUCAGCUGUCUGGGCAUCGUCAUC---GCAUGGGGAUCGGCGGCAUCUUGCUGCCGAGCUGUGCCAGUGUCCACUGGUCGGU- ..((((.....((.(((((....)))))))....)))).((.((((.---..)))).))((((((((.....))))))))((((.((((((....))))))))))- ( -40.40, z-score = -0.34, R) >droAna3.scaffold_12916 3422541 86 + 16180835 GGCCCUGUCAAAG--------UCU----GUGCCCUG---UCGUCG-----GGCUCGUCAUCGGCGGCAUCUUGCUGAAAAGUUUCUCUGGUGUUCCUUCAAUGUUC .(((((.....))--------...----(.((((..---.....)-----))).)......)))((((((..(((....)))......))))))............ ( -17.40, z-score = 0.73, R) >dp4.chr4_group3 3389047 81 - 11692001 GGUACUGUCAAAG--------UCUUCAGCUGUCGUGG--UCGUCGUC---GUAUGGGGAUCGUCGGCAUCUUGCAGUCCAGU--GGCCAGUGUCCA---------- ((((((((((..(--------.((.(((.((((((((--((.((((.---..)))).))))).)))))..))).)).)...)--)).))))).)).---------- ( -25.50, z-score = -1.49, R) >consensus GGGCCUGUCAAAGUCUUGAGCUCUUCAGCUGUCUUGGUAUCGUCAUC___GUAUGGGGAUCGGCGGCAUCUUGCUGCCGAGCUUUGCCAGUGUCCACUGGUC____ (((((((.((((((.......((((((..........................))))))((((((((.....)))))))))))))).))).))))........... (-22.16 = -24.56 + 2.39)

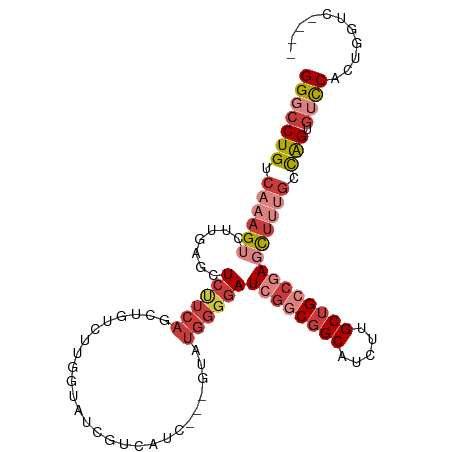
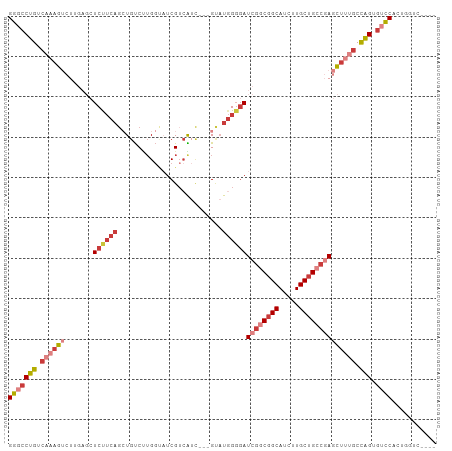
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:37 2011