
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,634,264 – 12,634,358 |
| Length | 94 |
| Max. P | 0.825037 |

| Location | 12,634,264 – 12,634,358 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.21 |
| Shannon entropy | 0.61306 |
| G+C content | 0.45465 |
| Mean single sequence MFE | -14.46 |
| Consensus MFE | -7.67 |
| Energy contribution | -7.15 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.825037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
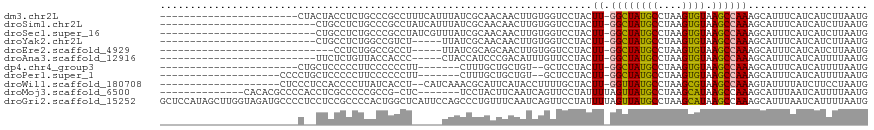
>dm3.chr2L 12634264 94 + 23011544 -----------------------CUACUACCUCUGCCCGCCUUUCAUUUAUCGCAACAACUUGUGGUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG -----------------------..........(((.............(((((((....))))))).....((-((((((((....)))).)))))).)))................ ( -15.00, z-score = -0.72, R) >droSim1.chr2L 12441583 91 + 22036055 --------------------------CUGCCUCUGCCCGCCUAUCAUUUAUCGCAACAACUUGUGGUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG --------------------------.......(((.............(((((((....))))))).....((-((((((((....)))).)))))).)))................ ( -15.00, z-score = -0.20, R) >droSec1.super_16 818932 91 + 1878335 --------------------------CUGCCUCUGCCCGCCUAUCGUUUAUCGCAACAACUUGUGGUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG --------------------------.......(((.........((..(((((((....)))))))...))((-((((((((....)))).)))))).)))................ ( -15.90, z-score = -0.42, R) >droYak2.chr2L 9070923 86 + 22324452 --------------------------CUGCCUCUGGCCGUCU-----UUAUCGCAACAACUUGUGGUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG --------------------------.(((...((((.....-----..(((((((....)))))))..(((((-(((...))).)))))...))))..)))................ ( -17.80, z-score = -0.98, R) >droEre2.scaffold_4929 13851214 83 - 26641161 -----------------------------CCUCUGGCCGCCU-----UUAUCGCAGCAACUUGUGGUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG -----------------------------.((.((((.....-----..(((((((....)))))))..(((((-(((...))).)))))...)))).)).................. ( -15.90, z-score = 0.24, R) >droAna3.scaffold_12916 3411197 86 - 16180835 --------------------------UUCUCUGUUACCACCC-----CUACCAUCCCGACAUUUGUUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUUUUAAUG --------------------------.....((((.......-----..........(((....))).....((-((((((((....)))).))))))))))................ ( -9.60, z-score = -0.57, R) >dp4.chr4_group3 3373969 85 + 11692001 -----------------------CUGCUCCCCCUUCCCCCCUU-------CUUUGCUGCUGU--GCUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUUUUAAUG -----------------------....................-------...((((..((.--(((..(((((-(((...))).)))))..))))).))))................ ( -16.40, z-score = -2.61, R) >droPer1.super_1 4850952 88 + 10282868 --------------------CCCCUGCUCCCCCUUCCCCCCUU-------CUUUGCUGCUGU--GCUCCUACUU-GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUUUUAAUG --------------------.......................-------...((((..((.--(((..(((((-(((...))).)))))..))))).))))................ ( -16.40, z-score = -2.89, R) >droWil1.scaffold_180708 2089964 95 - 12563649 --------------------CUCCCUCCACCCCUUAUCACCU--CAUCAAACGCAUUCAUACCUUUUGCUACUU-GGUUAUGCCUAAGCGUAAGCCAAAGUAUUUUAUCUUCCUAAUG --------------------......................--........(((...........)))(((((-((((((((....)))).)))).)))))................ ( -12.00, z-score = -2.36, R) >droMoj3.scaffold_6500 4023170 96 + 32352404 --------------CACACGCCCCACCUCGCCCCCGCCG-CUC-------UCCUACUUCAAUCAGUUCCUAUUUUAGUUAUGCCUAAGCAUAAGCCAAAGCAUUUAAUCAUUUUAAUG --------------...............((....((..-...-------....(((......)))...........((((((....))))))))....))................. ( -6.40, z-score = 0.15, R) >droGri2.scaffold_15252 15335678 118 + 17193109 GCUCCAUAGCUUGGUAGAUGCCCCUCCUCCGCCCCACUGGCUCAUUCCAGCCCUGUUUCAAUCAGUUCCUAUUUUAGUUAUGCCUAAGCAUAAGCCAAAGCAUUUAAUCAUUUUAAUG ........(((((((..((((.................((((......))))(((.......)))......................))))..))).))))................. ( -18.70, z-score = -0.42, R) >consensus _______________________C__CUCCCUCUGCCCGCCU_____UUAUCGCAACAACUUGUGGUCCUACUU_GGCUAUGCCUAAGUGUAAGCCAAAGCAUUUCAUCAUCUUAAUG .......................................................................(((.((((((((....)))).)))).))).................. ( -7.67 = -7.15 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:36 2011