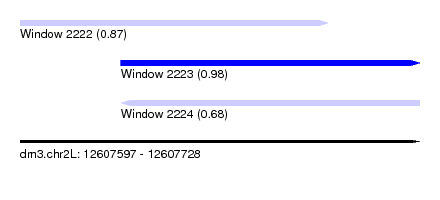
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,607,597 – 12,607,728 |
| Length | 131 |
| Max. P | 0.979070 |

| Location | 12,607,597 – 12,607,698 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.70462 |
| G+C content | 0.42595 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -9.79 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12607597 101 + 23011544 CCAGACUUCUUAG--GGCUGUUUUC-CGGGCUCAAUCUUAUGCAAAAUCACCUUAAGG---GAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAU------ .(((((..(((.(--((((......-..)))))..(((((((((((.(..((....))---..).-)))))))))))))).))))).....(((((.....)))))..------ ( -32.00, z-score = -1.92, R) >droSim1.chr2L 12412705 100 + 22036055 CCAGACUUCUUAG--GGCUGUUUUC-CGG-CUCAAUCUUAUGCAAAAUCACCUUAAGG---GAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAU------ .(((((..(((.(--(((((.....-)))-)))..(((((((((((.(..((....))---..).-)))))))))))))).))))).....(((((.....)))))..------ ( -34.10, z-score = -2.85, R) >droSec1.super_16 792463 100 + 1878335 CCAGACUUCUUAG--GGCUGUUUUC-CGG-CUCAAUCUUAUGCAAAAUCACCUUAAGG---GAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAU------ .(((((..(((.(--(((((.....-)))-)))..(((((((((((.(..((....))---..).-)))))))))))))).))))).....(((((.....)))))..------ ( -34.10, z-score = -2.85, R) >droYak2.chr2L 9043143 100 + 22324452 CCAGACUUCUUAG--GGUUGUUUUC-CAG-CUCAAUCUUAUGCAAAAUCACCUUAAGG---GAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAU------ .(((((..(((.(--(((((.....-)))-)))..(((((((((((.(..((....))---..).-)))))))))))))).))))).....(((((.....)))))..------ ( -32.40, z-score = -2.61, R) >droEre2.scaffold_4929 13824651 101 - 26641161 CCAGACUUCUUAG--GGUUGUUUUC-CAG-CUCAAUCUUAUGCAAAAUCACCUUAAGG---GAAAAUUUGCAUGGGAAAGUGUUUGUGAGGGUUCCUAAAUGGGACAU------ .(((((..(((.(--(((((.....-)))-)))..(((((((((((.((.((....))---))...)))))))))))))).))))).....(((((.....)))))..------ ( -31.60, z-score = -2.26, R) >droAna3.scaffold_12916 3383921 93 - 16180835 CCAGACUUCUUAG--GGUUGUUUUC-CAU-UUCAAUCUUAUGCAAAAUCACCUUAAGG---AGAA-UUUGCAUGGGAAAGUGUUUGUGCGGAUGAGAACAU------------- .....((((((((--(((.(((((.-(((-.........))).))))).)))))))))---)).(-(((((((..(......)..))))))))........------------- ( -26.10, z-score = -1.99, R) >dp4.chr4_group3 3345774 89 + 11692001 CCAGACUUCUUAG--GGUUGGUUCGGCCUCUGAAAUCUUAUGCAAAAUCAUCUUUAGG---GGACAUUUGCAUGGGAGAGUGCCUGAGAAAGUG-------------------- ....(((((((((--(.....(((((...))))).(((((((((((.((..(....).---.))..))))))))))).....)))))).)))).-------------------- ( -25.30, z-score = -0.68, R) >droPer1.super_1 4822790 89 + 10282868 CCAGACUUCUUAG--GGUUGGUUCGGCCUCUGAAAUCUUAUGCAAAAUCAUCUUUAGG---GGACAUUUGCAUGGGAGAGUGCCUGAGAAAGUG-------------------- ....(((((((((--(.....(((((...))))).(((((((((((.((..(....).---.))..))))))))))).....)))))).)))).-------------------- ( -25.30, z-score = -0.68, R) >droWil1.scaffold_180708 10445479 93 + 12563649 CCAGACUUCUUAGCCGGGUCUUUAGUCUCUAAGAACCUUAUGCAAAAUCAUUCAAGGGAGUGAAAAUUUGCAUGAGAAAGUGUUUUUGAGGUU--------------------- ...((((((..(((((((.(....).))).......((((((((((.((((((....))))))...))))))))))...).)))...))))))--------------------- ( -25.50, z-score = -1.79, R) >droVir3.scaffold_13246 779992 95 - 2672878 CCAGCCUGCUUAG--GACUCUUUAGGCC-----AAUCUUAUGCAAAAAAUCAUUGAAU------CAUUUGCAUGGGAAAGUGUUUCCCGAU--GCGCUCAAUCG----GAAAAG ...(((((...((--....)).))))).-----..(((((((((((..((......))------..))))))))))).........(((((--.......))))----)..... ( -21.90, z-score = -0.45, R) >droMoj3.scaffold_6500 4058370 93 + 32352404 CCAGAGUUCUUAG--CCGUCUUUAGGCC-----AAUCUUAUGCAAAAU--CCUUGAAU------AAUUUGCAUGGGAAAGUGUUUGGAAAU--GCAUUGAAGUG----GAAGGG (((((...(((.(--((.......))).-----..(((((((((((..--........------..))))))))))))))..)))))...(--.(((....)))----.).... ( -19.80, z-score = 0.35, R) >droGri2.scaffold_15252 15373964 99 + 17193109 CUCGAC-ACUUUC--CUAGCCUUGGGCC-----AAUCUUAUGCAAAAU-CCUUGGAAU------CAUUUGCAUGGGAAAGUGUUUUCCGCCUAACAUUCAAUCGAUGUGAAAAG ...(((-((((((--((((((...))).-----.......((((((.(-(....))..------..)))))))))))))))))).........(((((.....)))))...... ( -24.90, z-score = -1.78, R) >consensus CCAGACUUCUUAG__GGUUGUUUUC_CCG_CUCAAUCUUAUGCAAAAUCACCUUAAGG___GAAAAUUUGCAUGGGAAAGUGUUUGUGAGGGUGCCUAAAUGGG__________ ...................................(((((((((((....................)))))))))))..................................... ( -9.79 = -9.80 + 0.01)

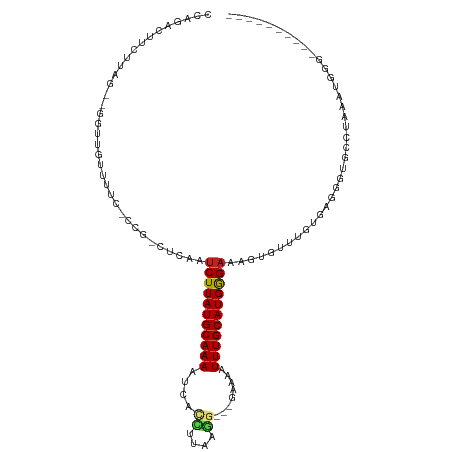
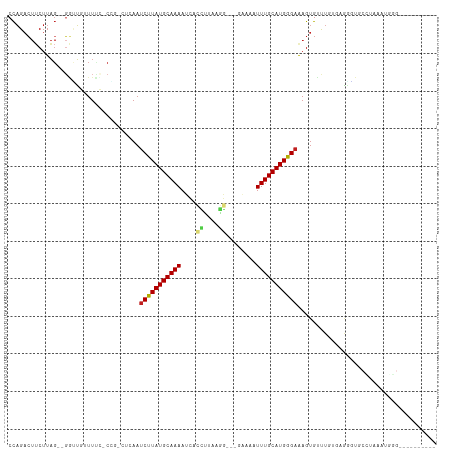
| Location | 12,607,630 – 12,607,728 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 61.55 |
| Shannon entropy | 0.80637 |
| G+C content | 0.40292 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.54 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12607630 98 + 23011544 CUUAUGCAAAAUCACCUUAAGGGAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAUCUUUCAAGGGUAUUGUUCUAUGUACUUAUU---- ((((((((((.(..((....))..).-)))))))))).........((((((....))..((((((((.(((....)))...)))))))))))).....---- ( -24.50, z-score = -1.51, R) >droSim1.chr2L 12412737 98 + 22036055 CUUAUGCAAAAUCACCUUAAGGGAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAUCUUUCAAGGGUAUUGUUCUAUGUCUUUGUU---- ((((((((((.(..((....))..).-))))))))))...............(((((.....)))))......(((((..((......))..)))))..---- ( -24.70, z-score = -1.41, R) >droSec1.super_16 792495 98 + 1878335 CUUAUGCAAAAUCACCUUAAGGGAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAUCUUUCAAGGCUAUGGUUCUAUGUACUUAUU---- ((((((((((.(..((....))..).-)))))))))).........(((((.(((((.....))))).......(..((....))..).))))).....---- ( -23.30, z-score = -0.91, R) >droYak2.chr2L 9043175 98 + 22324452 CUUAUGCAAAAUCACCUUAAGGGAAA-UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAUCUUUCAAGGGCAUUGUUCUAUGGAUUUGUU---- ((((((((((.(..((....))..).-))))))))))...((.((((.....(((((.....)))))......)))).))...................---- ( -25.00, z-score = -1.06, R) >droEre2.scaffold_4929 13824683 99 - 26641161 CUUAUGCAAAAUCACCUUAAGGGAAAAUUUGCAUGGGAAAGUGUUUGUGAGGGUUCCUAAAUGGGACAUCUUUCAAGGGUAUUCUUCUACGGAUUUACU---- ((((((((((.((.((....))))...))))))))))......((((.(((((((((.....))))).)))).))))((((....((....))..))))---- ( -26.90, z-score = -2.09, R) >droAna3.scaffold_12916 3383953 98 - 16180835 CUUAUGCAAAAUCACCUUAAGGAGA-AUUUGCAUGGGAAAGUGUUUGUGCGGAUGAGAACAUGC----UUAGGGGCUAUUCUCAUCCUGUAGGGGGAUUAGAU ((((((((((.((.((....)).))-.))))))))))..(((.(((.((((((((((((...((----(....)))..))))))))).))).))).))).... ( -36.20, z-score = -4.41, R) >dp4.chr4_group3 3345808 79 + 11692001 CUUAUGCAAAAUCAUCUUUAGGGGACAUUUGCAUGGGAGAGUGCCUGAGAAAGUGUUUCCAUGG----CUAAUGGCAGAUUUU-------------------- .......((((((.......((..((((((.(.((((......)))).).))))))..))...(----(.....)).))))))-------------------- ( -19.30, z-score = -0.31, R) >droPer1.super_1 4822824 79 + 10282868 CUUAUGCAAAAUCAUCUUUAGGGGACAUUUGCAUGGGAGAGUGCCUGAGAAAGUGUUUCCAUGG----CUAAUGGGAGAUUUU-------------------- ((((((((((.((..(....)..))..))))))))))............(((((.(((((((..----...))))))))))))-------------------- ( -17.60, z-score = 0.13, R) >droVir3.scaffold_13246 780021 87 - 2672878 CUUAUGCAAAAAAUCAUUGAAUCA---UUUGCAUGGGAAAGUGUUUCCCGAU--GCGCUCAAUCGGAA-AAGAAGCGGCCGCGCUUCAAGGUU---------- ((((((((((..((......))..---))))))))))..........(((((--.......)))))..-..((((((....))))))......---------- ( -23.80, z-score = -0.88, R) >droGri2.scaffold_15252 15373992 89 + 17193109 CUUAUGCAAAAU-CCUUGGAAUCA---UUUGCAUGGGAAAGUGUUUUCCGCCUAACAUUCAAUCGAUGUGAAAAGAGACCGAGAUUUAAGACU---------- ........((((-(.((((..((.---((..(((.((..((((((........))))))...)).)))..))..))..)))))))))......---------- ( -17.60, z-score = -0.47, R) >consensus CUUAUGCAAAAUCACCUUAAGGGAAA_UUUGCAUGGGAAAGUGUUUGUGCGGGUUCCUAAAUGGGACAUCAUUCAAGGGUAUUGUUCUAUGUA_UU_UU____ ((((((((((....((....)).....)))))))))).................................................................. (-10.52 = -10.54 + 0.02)
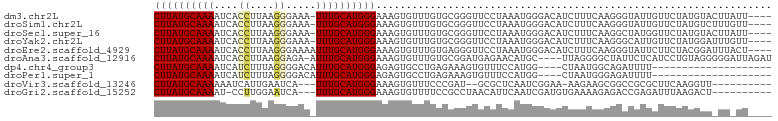
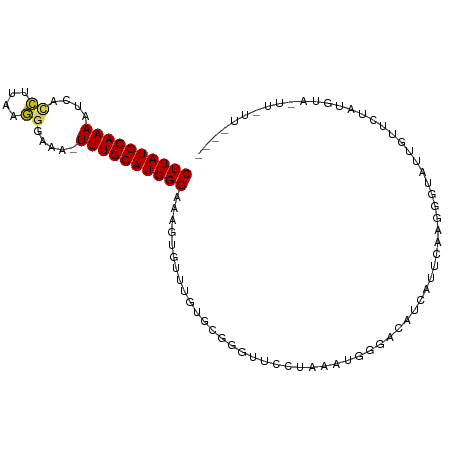
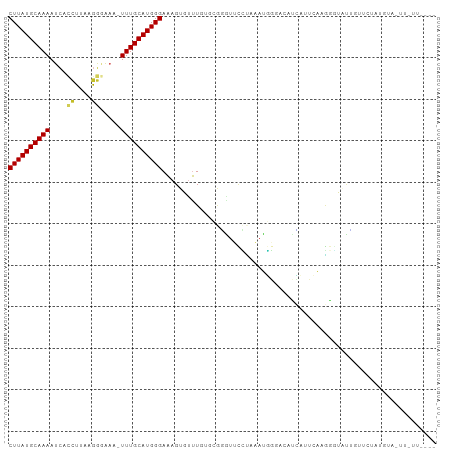
| Location | 12,607,630 – 12,607,728 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 61.55 |
| Shannon entropy | 0.80637 |
| G+C content | 0.40292 |
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -4.56 |
| Energy contribution | -4.56 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12607630 98 - 23011544 ----AAUAAGUACAUAGAACAAUACCCUUGAAAGAUGUCCCAUUUAGGAACCCGCACAAACACUUUCCCAUGCAAA-UUUCCCUUAAGGUGAUUUUGCAUAAG ----.........................(((((.(((........((...))......))))))))..(((((((-.(..((....))..).)))))))... ( -14.04, z-score = -0.86, R) >droSim1.chr2L 12412737 98 - 22036055 ----AACAAAGACAUAGAACAAUACCCUUGAAAGAUGUCCCAUUUAGGAACCCGCACAAACACUUUCCCAUGCAAA-UUUCCCUUAAGGUGAUUUUGCAUAAG ----......(((((....(((.....)))....))))).......((((..............)))).(((((((-.(..((....))..).)))))))... ( -15.64, z-score = -1.59, R) >droSec1.super_16 792495 98 - 1878335 ----AAUAAGUACAUAGAACCAUAGCCUUGAAAGAUGUCCCAUUUAGGAACCCGCACAAACACUUUCCCAUGCAAA-UUUCCCUUAAGGUGAUUUUGCAUAAG ----.........................(((((.(((........((...))......))))))))..(((((((-.(..((....))..).)))))))... ( -14.04, z-score = -0.57, R) >droYak2.chr2L 9043175 98 - 22324452 ----AACAAAUCCAUAGAACAAUGCCCUUGAAAGAUGUCCCAUUUAGGAACCCGCACAAACACUUUCCCAUGCAAA-UUUCCCUUAAGGUGAUUUUGCAUAAG ----.........................(((((.(((........((...))......))))))))..(((((((-.(..((....))..).)))))))... ( -14.04, z-score = -0.56, R) >droEre2.scaffold_4929 13824683 99 + 26641161 ----AGUAAAUCCGUAGAAGAAUACCCUUGAAAGAUGUCCCAUUUAGGAACCCUCACAAACACUUUCCCAUGCAAAUUUUCCCUUAAGGUGAUUUUGCAUAAG ----.........(((......)))....(((((.(((.......(((...))).....))))))))..(((((((...((((....)).)).)))))))... ( -14.30, z-score = -0.58, R) >droAna3.scaffold_12916 3383953 98 + 16180835 AUCUAAUCCCCCUACAGGAUGAGAAUAGCCCCUAA----GCAUGUUCUCAUCCGCACAAACACUUUCCCAUGCAAAU-UCUCCUUAAGGUGAUUUUGCAUAAG ................(((((((((((((......----)).)))))))))))................(((((((.-((.((....)).)).)))))))... ( -30.20, z-score = -6.31, R) >dp4.chr4_group3 3345808 79 - 11692001 --------------------AAAAUCUGCCAUUAG----CCAUGGAAACACUUUCUCAGGCACUCUCCCAUGCAAAUGUCCCCUAAAGAUGAUUUUGCAUAAG --------------------......((((.....----...((....))........)))).......(((((((.(((..(....)..))))))))))... ( -13.69, z-score = -0.81, R) >droPer1.super_1 4822824 79 - 10282868 --------------------AAAAUCUCCCAUUAG----CCAUGGAAACACUUUCUCAGGCACUCUCCCAUGCAAAUGUCCCCUAAAGAUGAUUUUGCAUAAG --------------------..............(----((.(((((.....))).)))))........(((((((.(((..(....)..))))))))))... ( -13.10, z-score = -1.15, R) >droVir3.scaffold_13246 780021 87 + 2672878 ----------AACCUUGAAGCGCGGCCGCUUCUU-UUCCGAUUGAGCGC--AUCGGGAAACACUUUCCCAUGCAAA---UGAUUCAAUGAUUUUUUGCAUAAG ----------......((((((....))))))((-(((((((.......--))))))))).........(((((((---.((((....)))).)))))))... ( -23.00, z-score = -1.59, R) >droGri2.scaffold_15252 15373992 89 - 17193109 ----------AGUCUUAAAUCUCGGUCUCUUUUCACAUCGAUUGAAUGUUAGGCGGAAAACACUUUCCCAUGCAAA---UGAUUCCAAGG-AUUUUGCAUAAG ----------.((((...((.((((((............))))))))...))))(((((....))))).(((((((---....((....)-).)))))))... ( -18.30, z-score = -1.07, R) >consensus ____AA_AA_UACAUAGAACAAUACCCUCGAAAGAUGUCCCAUGUAGGAACCCGCACAAACACUUUCCCAUGCAAA_UUUCCCUUAAGGUGAUUUUGCAUAAG .....................................................................(((((((...((......))....)))))))... ( -4.56 = -4.56 + 0.00)
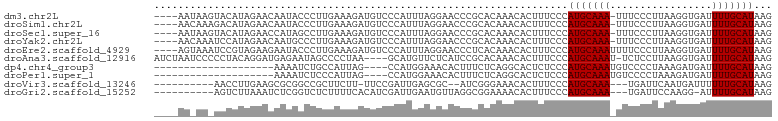
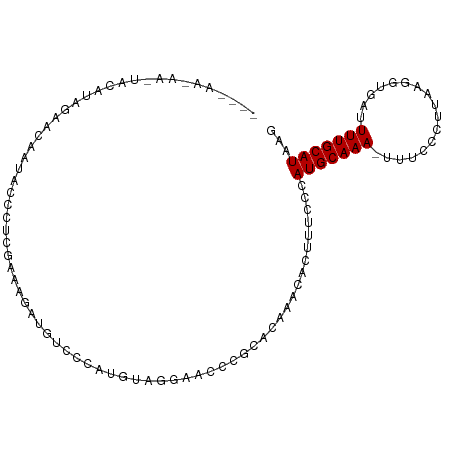
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:33 2011