
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,606,654 – 12,606,745 |
| Length | 91 |
| Max. P | 0.621552 |

| Location | 12,606,654 – 12,606,745 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.72 |
| Shannon entropy | 0.50709 |
| G+C content | 0.33147 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -9.71 |
| Energy contribution | -9.24 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
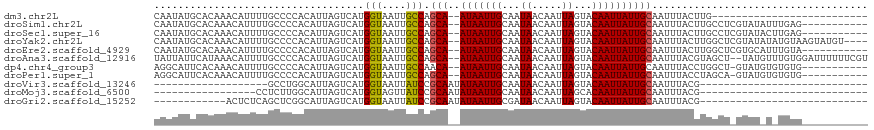
>dm3.chr2L 12606654 91 - 23011544 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUG-------------------------- (((.(((......))).)))..............((((....))))(((--(((((((...((.....))...))))))))))..........-------------------------- ( -14.30, z-score = -0.22, R) >droSim1.chr2L 12411759 106 - 22036055 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUGCCUCGUAUAUUUGAG----------- .(((((((..........................((((....))))(((--(.(((((((((((............)))))))))))...))))...)))))))....----------- ( -20.70, z-score = -1.43, R) >droSec1.super_16 791508 106 - 1878335 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUGCCUCGUAUACUUGAG----------- ((((((((..........................((((....))))(((--(.(((((((((((............)))))))))))...))))...))))).)))..----------- ( -20.20, z-score = -1.31, R) >droYak2.chr2L 9042135 113 - 22324452 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUGGCUCGUAUAUAUGUAAGUAUGU---- ..((((((.........(((((..((....))...)))))..(((((..--..(((((((((((............)))))))))))...)))))..))))))............---- ( -23.30, z-score = -0.66, R) >droEre2.scaffold_4929 13823723 106 + 26641161 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUGGCUCGUGCAUUUGUA----------- (((.((((((.......(((((..((....))...)))))..(((((..--..(((((((((((............)))))))))))...)))))..)))))))))..----------- ( -27.40, z-score = -2.36, R) >droAna3.scaffold_12916 3383061 115 + 16180835 UAUUAUUCAUAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACGUAGCU--UAUGUUUGUGGAUUUUUUCGU ....((((((((((((...((..(..........((((....))))(((--(((((((...((.....))...)))))))))).......)..)).--.))))))))))))........ ( -28.10, z-score = -3.22, R) >dp4.chr4_group3 3344402 105 - 11692001 AGGCAUUCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAACA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACCUGGCU-GUAUGUGUGUG----------- .((((.............)))).(((((((((((.((((((((....))--))(((((((((((............))))))))))))))))))))-).)))))....----------- ( -28.22, z-score = -2.10, R) >droPer1.super_1 4821425 105 - 10282868 AGGCAUUCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA--AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACCUAGCA-GUAUGUGUGUG----------- .......((((.((((.((((.............((((....))))(((--(((((((...((.....))...))))))))))..........)))-).)))).))))----------- ( -24.50, z-score = -1.42, R) >droVir3.scaffold_13246 1891713 72 - 2672878 -------------------GCCUGGCAUUAGUCAUGGUAAUUAUCCGCAAUAUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACG---------------------------- -------------------(((((((....)))).)))...............(((((((((((............)))))))))))....---------------------------- ( -14.20, z-score = -1.62, R) >droMoj3.scaffold_6500 28292783 74 + 32352404 -----------------CCUCUUGGCAUUAGUCAUGGUAGUUAUCCGCAAUAUAAUUGCAAUAACAAUUAGCACAAUUAUUGCAAUUUACG---------------------------- -----------------((...((((....)))).)).........((((((....((((((....))).)))....))))))........---------------------------- ( -12.40, z-score = -1.04, R) >droGri2.scaffold_15252 1817947 79 + 17193109 ------------ACUCUCAGCUCGGCAUUAGUCAUGGUAAUUAUCCGCAAUAUAAUUGCGAUAACAAUUAGUACAAUUAUUGCAAUUUACG---------------------------- ------------............(((.((((..((.(((((...((((((...)))))).....))))).))..)))).)))........---------------------------- ( -12.50, z-score = -1.00, R) >consensus CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCA__AUAAUUGCAAUAACAAUUAGUACAAUUAUUGCAAUUUACUUGGCU_GUAU_U_UGUG___________ ...................................(((....)))........(((((((((((............)))))))))))................................ ( -9.71 = -9.24 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:31 2011