| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,574,330 – 12,574,424 |
| Length | 94 |
| Max. P | 0.622960 |

| Location | 12,574,330 – 12,574,424 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Shannon entropy | 0.39795 |
| G+C content | 0.40804 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
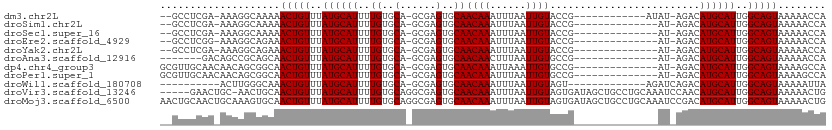
>dm3.chr2L 12574330 94 - 23011544 --GCCUCGA-AAAGGCAAAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACCG------------AUAU-AGACAUGCAUUGGCAGUAAAAACCA --((((...-..))))....(((((..((((((((((((..-.((.((((((.........))))))))------------.)))-))).))))))..)))))........ ( -27.80, z-score = -3.03, R) >droSim1.chr2L 12376472 92 - 22036055 --GCCUCGA-AAAGGCAAAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACCG--------------AU-AGACAUGCAUUGGCAGUAAAAACCA --((((...-..))))....(((((..(((((((((((...-.((.((((((.........))))))))--------------))-))).))))))..)))))........ ( -25.00, z-score = -2.05, R) >droSec1.super_16 757435 92 - 1878335 --GCCUCGA-AAAGGCAAAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACCG--------------AU-AGACAUGCAUUGGCAGUAAAAACCA --((((...-..))))....(((((..(((((((((((...-.((.((((((.........))))))))--------------))-))).))))))..)))))........ ( -25.00, z-score = -2.05, R) >droEre2.scaffold_4929 13790348 92 + 26641161 --GCCUCGG-AAAGGCAGAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACCG--------------AU-AGACAUGCAUUGGCAGUAAAAACCA --((((...-..))))....(((((..(((((((((((...-.((.((((((.........))))))))--------------))-))).))))))..)))))........ ( -25.00, z-score = -1.60, R) >droYak2.chr2L 9008362 92 - 22324452 --GCCUCGA-AAAGGCAGAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACCG--------------AU-AGACAUGCAUUGGCAGUAAAAACCA --((((...-..))))....(((((..(((((((((((...-.((.((((((.........))))))))--------------))-))).))))))..)))))........ ( -25.00, z-score = -1.91, R) >droAna3.scaffold_12916 3351123 88 + 16180835 -------GACAGCCGCAGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAACUUUAAUUGUGCCG--------------AU-AGACAUGCAUUGGCAGUAAAAACCA -------....((((..(((..(((((((((((...)))).-.((.(..(((.........)))..)))--------------))-))))))))..))))........... ( -21.20, z-score = 0.37, R) >dp4.chr4_group3 3298784 95 - 11692001 GCGUUGCAACAACAGCGGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUAAAUUGUGCCG--------------AU-AGACAUGCAUUGGCAGUAAAAGCCA ((((((...)))).))(((.(((((..(((((((((((...-.((.(..(((.........)))..)))--------------))-))).))))))..)))))....))). ( -26.50, z-score = 0.24, R) >droPer1.super_1 4779372 95 - 10282868 GCGUUGCAACAACAGCGGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUGCCG--------------AU-AGACAUGCAUUGGCAGUAAAAGCCA ((((((...)))).))(((.(((((..(((((((((((...-.((.(..(((.........)))..)))--------------))-))).))))))..)))))....))). ( -26.50, z-score = 0.28, R) >droWil1.scaffold_180708 10373875 87 - 12563649 ----------ACUUGGGCAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUAGU-------------AGAUCAGACAUGCAUUGGCAGUAAAAAUUA ----------...((.(((((.(((....))).))))).))-((.((((((((((......)))).((-------------.......)).)))))).))........... ( -19.40, z-score = -0.06, R) >droVir3.scaffold_13246 1842737 105 - 2672878 -----GAACUGC-AACUGCAACUGUUUAUGCAUUUUGUGCAGGCGAGUGCAACAAAUUUAAUUGUAGUGAUAGCUGCCUGCAAAUCCAACAUGCAUUGGCAGUAAAAACUG -----....(((-....)))(((((..((((((.((((((((((.(((.((((((......))))..))...))))))))))....))).))))))..)))))........ ( -34.10, z-score = -2.26, R) >droMoj3.scaffold_6500 28246684 111 + 32352404 AACUGCAACUGCAAAGUGCAACUGUUUAUGCAUUUUGUGCAGGCGAGUGCAACAAAUUUAAUUGUAGUGAUAGCUGCCUGCAAAUCCGACAUGCAUUGGCAGUAAAAACUG ...((((.((....))))))(((((..((((((.((((((((((.(((.((((((......))))..))...))))))))))....))).))))))..)))))........ ( -36.00, z-score = -1.89, R) >consensus __GCCUCGA_AAAGGCAGAAACUGUUUAUGCAUUUUGUGCA_GCGAGUGCAACAAAUUUAAUUGUACCG______________AU_AGACAUGCAUUGGCAGUAAAAACCA ....................(((((..((((((..((..(......)..))((((......)))).........................))))))..)))))........ (-17.78 = -17.78 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:29 2011