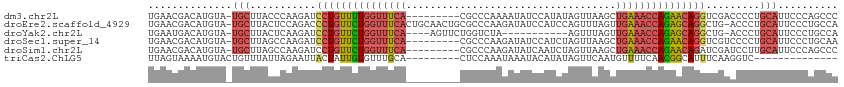
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,154,456 – 1,154,561 |
| Length | 105 |
| Max. P | 0.999828 |

| Location | 1,154,456 – 1,154,561 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.94 |
| Shannon entropy | 0.59936 |
| G+C content | 0.43934 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.82 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
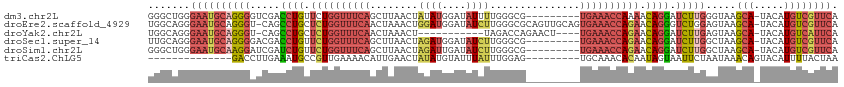
>dm3.chr2L 1154456 105 + 23011544 GGGCUGGGAAUGCAGGGGUCGACCUGUUCUGGUUUCAGCUUAACUAUAUGGAUAUUUUGGGCG---------UGAAACCAAAACAGGAUCUUGGGUAAGCA-UACAUGUCGUUCA .(((...(.((((...(.((((((((((.((((((((((((((..(((....))).)))))).---------)))))))).))))))...)))).)..)))-).)..)))..... ( -33.00, z-score = -2.06, R) >droEre2.scaffold_4929 1198893 113 + 26641161 UGGCAGGGAAUGCAGGGU-CAGCCUGCUCUGGUUUCAACUAAACUGGAUGGAUAUCUUGGGCGCAGUUGCAGUGAAACCAGAACAGGGUCUGGAGUAAGCA-UACAUGUCGUUCA .((((..(.((((....(-(((((((.((((((((((....((((((..((....))....).)))))....)))))))))).))))..)))).....)))-).).))))..... ( -39.30, z-score = -1.78, R) >droYak2.chr2L 1133193 98 + 22324452 UGGCAGGGAAUGCAGGGU-CAGCCUGCUCUGGUUUCAACUAAACU-----------UAGACCAGAACU----UGAAACCAGAACAGGAUCUUGAGUAAGCA-UACAUGUCAUUCA (((((..(.((((..(.(-((.((((.(((((((((((.......-----------...........)----)))))))))).))))....))).)..)))-).).))))).... ( -32.37, z-score = -2.63, R) >droSec1.super_14 1114462 105 + 2068291 UUGCAGGGAAUGCAGGGGACGACCUGUUCUGGUUUCAGCUUAACUAGAUGGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUCUUGGCUAAGCA-UACAUGUCGUUCA (((((.....))))).((((((((((((((((((((((((((...((((....))))))))).---------)))))))))))))))......((...)).-......)))))). ( -42.40, z-score = -4.60, R) >droSim1.chr2L 1126437 105 + 22036055 GGGCUGGGAAUGCAAGGAUCGAUCUGUUCUGGUUUCAGCUUAACUAGAUUGAUAUCUUGGGCG---------UGAAACCAGAACAGGAUCUUGGCUAAGCA-UACAUGUCGUUCA .(((((.(...((.((((((...(((((((((((((((((((...((((....))))))))).---------)))))))))))))))))))).))....).-..)).)))..... ( -40.00, z-score = -4.16, R) >triCas2.ChLG5 10857534 92 + 18847211 --------------GACCUUGAAAUGCCGUUGAAAACAUUGAACUAUAUGUAUUUAUUUGGAG---------UGCAAACACAAUAGUAAUUCUAAUAAACAGUACAUUUUACUAA --------------.....((((((((((.((((.((((........)))).))))..))).(---------((....))).......................))))))).... ( -10.90, z-score = 0.37, R) >consensus UGGCAGGGAAUGCAGGGGUCGACCUGCUCUGGUUUCAACUUAACUAGAUGGAUAUCUUGGGCG_________UGAAACCAGAACAGGAUCUUGAGUAAGCA_UACAUGUCGUUCA ............(((((.....((((.((((((((((...................................)))))))))).)))).)))))...................... (-12.65 = -13.82 + 1.17)
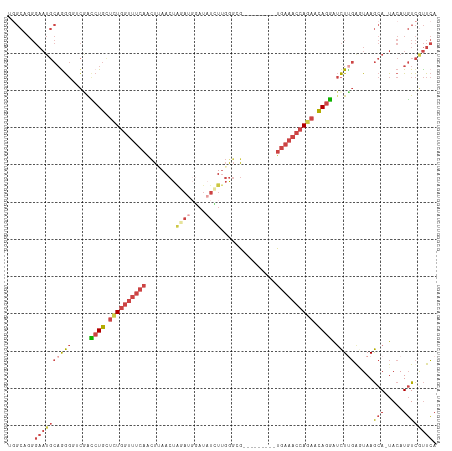
| Location | 1,154,456 – 1,154,561 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.94 |
| Shannon entropy | 0.59936 |
| G+C content | 0.43934 |
| Mean single sequence MFE | -28.55 |
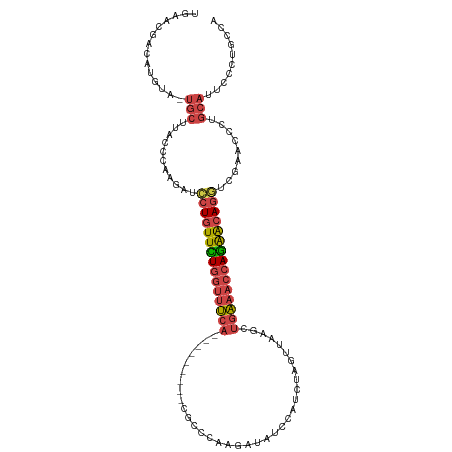
| Consensus MFE | -16.29 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
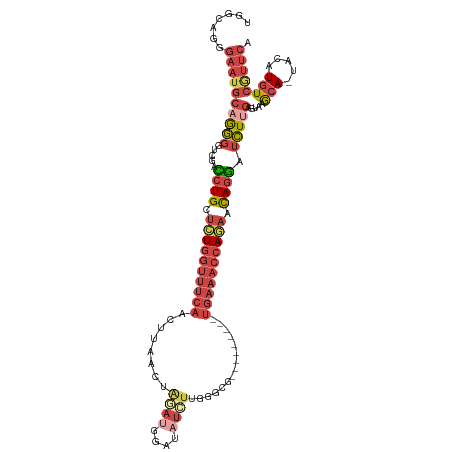
>dm3.chr2L 1154456 105 - 23011544 UGAACGACAUGUA-UGCUUACCCAAGAUCCUGUUUUGGUUUCA---------CGCCCAAAAUAUCCAUAUAGUUAAGCUGAAACCAGAACAGGUCGACCCCUGCAUUCCCAGCCC .........((.(-(((........((.(((((((((((((((---------.((......(((....))).....))))))))))))))))))).......))))...)).... ( -27.96, z-score = -4.21, R) >droEre2.scaffold_4929 1198893 113 - 26641161 UGAACGACAUGUA-UGCUUACUCCAGACCCUGUUCUGGUUUCACUGCAACUGCGCCCAAGAUAUCCAUCCAGUUUAGUUGAAACCAGAGCAGGCUG-ACCCUGCAUUCCCUGCCA .....(.((...(-(((......(((..((((((((((((((((((.(((((.................)))))))).))))))))))))))))))-.....))))....)).). ( -34.83, z-score = -3.55, R) >droYak2.chr2L 1133193 98 - 22324452 UGAAUGACAUGUA-UGCUUACUCAAGAUCCUGUUCUGGUUUCA----AGUUCUGGUCUA-----------AGUUUAGUUGAAACCAGAGCAGGCUG-ACCCUGCAUUCCCUGCCA .(((((.((.(((-....)))(((....(((((((((((((((----(.(.((......-----------))...).)))))))))))))))).))-)...)))))))....... ( -31.80, z-score = -2.94, R) >droSec1.super_14 1114462 105 - 2068291 UGAACGACAUGUA-UGCUUAGCCAAGAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGUCCCCUGCAUUCCCUGCAA .....(((.....-.((...))...((.(((((((((((((((---------.((.(.((((....)))).)....))))))))))))))))))))))...((((.....)))). ( -33.60, z-score = -4.60, R) >droSim1.chr2L 1126437 105 - 22036055 UGAACGACAUGUA-UGCUUAGCCAAGAUCCUGUUCUGGUUUCA---------CGCCCAAGAUAUCAAUCUAGUUAAGCUGAAACCAGAACAGAUCGAUCCUUGCAUUCCCAGCCC .........((.(-(((..((....((((.(((((((((((((---------.((.(.((((....)))).)....)))))))))))))))))))....)).))))...)).... ( -31.10, z-score = -4.44, R) >triCas2.ChLG5 10857534 92 - 18847211 UUAGUAAAAUGUACUGUUUAUUAGAAUUACUAUUGUGUUUGCA---------CUCCAAAUAAAUACAUAUAGUUCAAUGUUUUCAACGGCAUUUCAAGGUC-------------- ......((((((..((..((((.((((((....((((((((..---------.......))))))))..))))))))))....))...)))))).......-------------- ( -12.00, z-score = 0.24, R) >consensus UGAACGACAUGUA_UGCUUACCCAAGAUCCUGUUCUGGUUUCA_________CGCCCAAGAUAUCCAUCUAGUUAAGCUGAAACCAGAACAGGUCGAACCCUGCAUUCCCUGCCA ..............(((...........(((((((((((((((...................................))))))))))))))).........))).......... (-16.29 = -17.02 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:49 2011