
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,543,950 – 12,544,052 |
| Length | 102 |
| Max. P | 0.964908 |

| Location | 12,543,950 – 12,544,052 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Shannon entropy | 0.54235 |
| G+C content | 0.39904 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
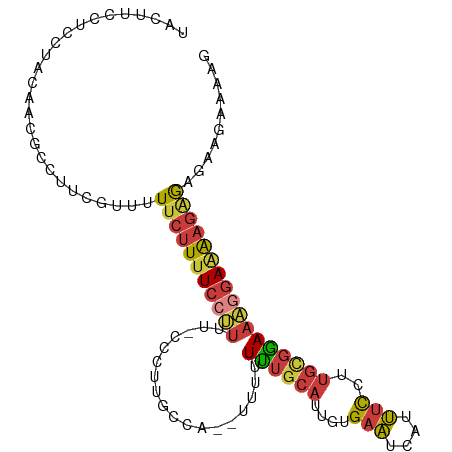
>dm3.chr2L 12543950 102 + 23011544 CUUUUCUUCGCUCUUUUCCUUUCCGCAAGGAAAUGAUUCACAAUGCAAAAAAAA-UGGCAAGGG-AAAAGGAAAAGAAAAACGAAGGCGUUGUAGGAGGAAGUA .((((((((..(((((((((((((....))).....(((.(..(((........-..)))..))-))))))))))))..((((....))))...)))))))).. ( -28.60, z-score = -2.26, R) >droSim1.chr2L 12353032 103 + 22036055 CUUUUCUUCGCUCUUUUCCUUUCCGCAAGGAAAUGAUUCACAAUGCAAAAAAAAAUGGCAAGGG-AAAAGGAAAAGAAAAACGAAGGCGUUGUAGGAGGAAGUA .((((((((..(((((((((((((....))).....(((.(..(((...........)))..))-))))))))))))..((((....))))...)))))))).. ( -28.50, z-score = -2.26, R) >droSec1.super_16 734047 101 + 1878335 CUUUUCUUCACUCUUUUCCUUUCCGCAAGGAAAUGAUUCACAAUGCAAAAAAA--UGGCAAGGG-AAAAGGAAAAGAAAAACGAAGGCGUUGUAGGAGGAAGUA .((((((((..(((((((((((((....))).....(((.(..(((.......--..)))..))-))))))))))))..((((....))))...)))))))).. ( -28.70, z-score = -2.64, R) >droYak2.chr2L 8983519 96 + 22324452 ---CUUUUCUCUCUUUUCCUUUUCGCAAGGAAAUGAUUCACAAUGCAAAAA----UGGCAAGGG-AAAAGGAAAAGAAAAACGAAGGCGUUGUAGGAGGAAGUA ---.((((((((((((((((((((.(...(((....)))....(((.....----..))).).)-))))))))))))..((((....))))....))))))).. ( -28.10, z-score = -2.86, R) >droEre2.scaffold_4929 13766691 101 - 26641161 CUUUUUCUCUCUCUUUUCCUUUCCGCAAGGAAAUGAUUCACAAUGCAAAAAAA--UGGCAAGGG-AAAAGGAAAAGAAAAACGAAGGCGUUAUAGGAGGAAGUA ..((((((((.(((((((((((((....))).....(((.(..(((.......--..)))..))-))))))))))))..((((....))))...)))))))).. ( -28.50, z-score = -2.60, R) >droAna3.scaffold_12916 3326880 97 - 16180835 -----AUAGCCCCAUCUCACUUCCACAAGGAAGUGAUUCACAAUGCAGAAGAG--UGUAAAGUGUAAAGGGAAAAGAAAAACGGAGGCGUUGUAGGAGGAAGCA -----...(((((...((((((((....))))))))..(((..((((......--))))..)))..................)).)))(((.........))). ( -23.80, z-score = -1.70, R) >droGri2.scaffold_15074 5541407 83 - 7742996 --AUUGCCGUUGUUCUUGUUGUUGUUGAAACCAUGAAUCAUGCUGCGAAAGC------------------GAUAACUAAAUUGGCCUAUUUGGCGAAGCAAGG- --............((((((((((((.....((((...))))..((....))------------------)))))).......(((.....)))..)))))).- ( -19.20, z-score = -0.49, R) >consensus CUUUUCUUCUCUCUUUUCCUUUCCGCAAGGAAAUGAUUCACAAUGCAAAAAAA__UGGCAAGGG_AAAAGGAAAAGAAAAACGAAGGCGUUGUAGGAGGAAGUA ..............((((((((((....)))))...((((((((((........................................))))))).)))))))).. (-12.05 = -12.00 + -0.05)



| Location | 12,543,950 – 12,544,052 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 73.17 |
| Shannon entropy | 0.54235 |
| G+C content | 0.39904 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -9.16 |
| Energy contribution | -9.72 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12543950 102 - 23011544 UACUUCCUCCUACAACGCCUUCGUUUUUCUUUUCCUUUU-CCCUUGCCA-UUUUUUUUGCAUUGUGAAUCAUUUCCUUGCGGAAAGGAAAAGAGCGAAGAAAAG ..................((((((...((((((((((((-(...(((.(-......).)))..(..(.........)..))))))))))))))))))))..... ( -23.70, z-score = -2.43, R) >droSim1.chr2L 12353032 103 - 22036055 UACUUCCUCCUACAACGCCUUCGUUUUUCUUUUCCUUUU-CCCUUGCCAUUUUUUUUUGCAUUGUGAAUCAUUUCCUUGCGGAAAGGAAAAGAGCGAAGAAAAG ..................((((((...((((((((((((-(.................(((..(.(((....)))).))))))))))))))))))))))..... ( -23.42, z-score = -2.25, R) >droSec1.super_16 734047 101 - 1878335 UACUUCCUCCUACAACGCCUUCGUUUUUCUUUUCCUUUU-CCCUUGCCA--UUUUUUUGCAUUGUGAAUCAUUUCCUUGCGGAAAGGAAAAGAGUGAAGAAAAG ..................(((((...(((((((((((((-(........--.......(((..(.(((....)))).))))))))))))))))))))))..... ( -20.66, z-score = -1.51, R) >droYak2.chr2L 8983519 96 - 22324452 UACUUCCUCCUACAACGCCUUCGUUUUUCUUUUCCUUUU-CCCUUGCCA----UUUUUGCAUUGUGAAUCAUUUCCUUGCGAAAAGGAAAAGAGAGAAAAG--- .......................((((((((((((((((-(...(((..----.....)))..(..(.........)..))))))))))))))))))....--- ( -21.30, z-score = -2.77, R) >droEre2.scaffold_4929 13766691 101 + 26641161 UACUUCCUCCUAUAACGCCUUCGUUUUUCUUUUCCUUUU-CCCUUGCCA--UUUUUUUGCAUUGUGAAUCAUUUCCUUGCGGAAAGGAAAAGAGAGAGAAAAAG .......................(((((((((..(((((-((.......--..((((((((..(.(((....)))).)))))))))))))))..))))))))). ( -23.50, z-score = -2.31, R) >droAna3.scaffold_12916 3326880 97 + 16180835 UGCUUCCUCCUACAACGCCUCCGUUUUUCUUUUCCCUUUACACUUUACA--CUCUUCUGCAUUGUGAAUCACUUCCUUGUGGAAGUGAGAUGGGGCUAU----- ................(((.(((((...............(((..(.((--......)).)..)))..((((((((....))))))))))))))))...----- ( -24.40, z-score = -3.16, R) >droGri2.scaffold_15074 5541407 83 + 7742996 -CCUUGCUUCGCCAAAUAGGCCAAUUUAGUUAUC------------------GCUUUCGCAGCAUGAUUCAUGGUUUCAACAACAACAAGAACAACGGCAAU-- -.........(((......((((....((((((.------------------(((.....)))))))))..))))(((...........)))....)))...-- ( -11.70, z-score = 0.70, R) >consensus UACUUCCUCCUACAACGCCUUCGUUUUUCUUUUCCUUUU_CCCUUGCCA__UUUUUUUGCAUUGUGAAUCAUUUCCUUGCGGAAAGGAAAAGAGAGAAGAAAAG ..........................(((((((((((..................((((((....(((....)))..))))))))))))))))).......... ( -9.16 = -9.72 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:27 2011