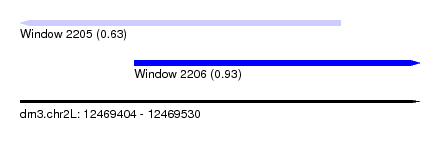
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,469,404 – 12,469,530 |
| Length | 126 |
| Max. P | 0.934781 |

| Location | 12,469,404 – 12,469,505 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Shannon entropy | 0.14171 |
| G+C content | 0.45936 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.628039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
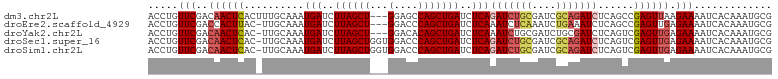
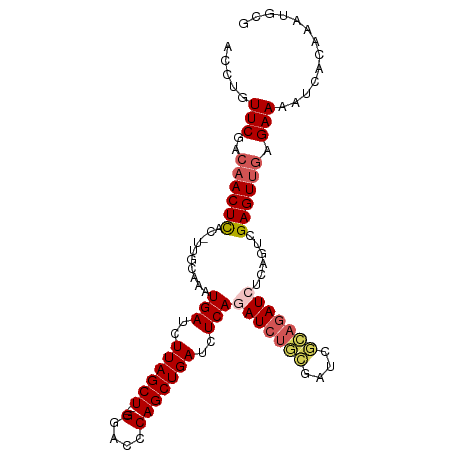
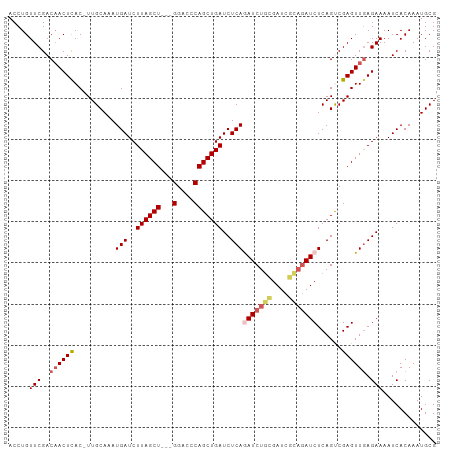
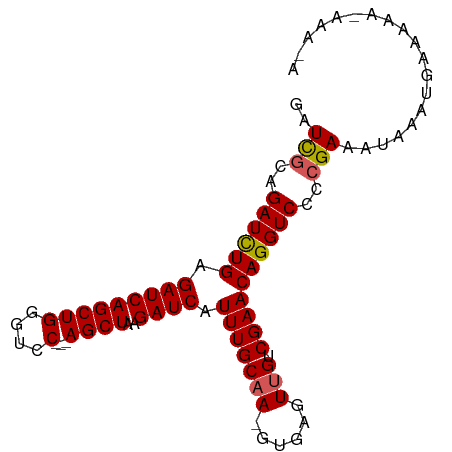
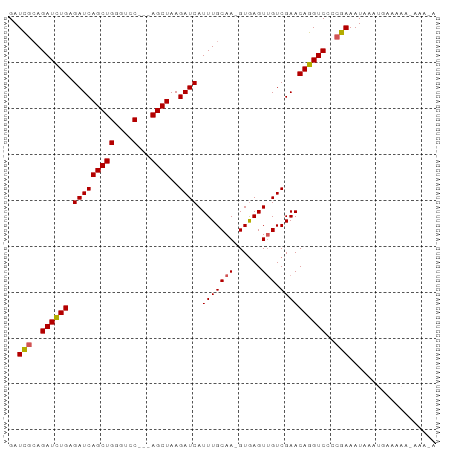
>dm3.chr2L 12469404 101 - 23011544 ACCUGUUCGACAACUCACUUUGCAAAUGAUCUUAGCU---GGAGCCAGCUGAUCUCAGAUCUGCGAUCGCAGAUCUCAGCCGAGUUAAGAAAAUCACAAAUGCG .....(((...(((((..........(((..((((((---(....)))))))..)))(((((((....)))))))......)))))..)))............. ( -30.40, z-score = -2.32, R) >droEre2.scaffold_4929 13691132 100 + 26641161 ACCUGUUCGACCACUUAC-UUGCAAAUGAUCUUAGCU---GGACCCAGCUGAUCUCAAAUCUCAAAUCUGAAAUCUCAGCCGAGUUGAGAAAAUCACAAAUGCG ..................-..(((..(((..((((((---(....)))))))..)))..((((((..((((....)))).....))))))..........))). ( -24.10, z-score = -2.72, R) >droYak2.chr2L 8906518 100 - 22324452 ACCUGUUCGACAACUCAC-UUGCAAAUGAUCUUAGCU---GGACACAGCUGAUCUCAAAUCUGCGAUCUGCGAUCUCAGUCGAGUUGAGAAAAUCACAAAUGCG .....(((..((((((..-.......(((..((((((---(....)))))))..)))...(((.((((...)))).)))..)))))).)))............. ( -26.50, z-score = -1.53, R) >droSec1.super_16 660254 103 - 1878335 ACCUGUUCGACAACUCAC-UUGCAAAUGAUCUUAGCUGGUGGACCCAGCUGAUCUCAGAUCUGCGAUCGCAGAUCUCAGUCGAGUUGAGAAAAUCACAAAUGCG .....(((..((((((((-(......(((..((((((((.....))))))))..)))(((((((....)))))))..))).)))))).)))............. ( -35.10, z-score = -2.97, R) >droSim1.chr2L 12278406 103 - 22036055 ACCUGUUCGACAACUCAC-UUGCAAAUGAUCUUAGCUGGUGGACCCAGCUGAUCUCAGAUCUGCGAUCGCAGAUCUCAGUCGAGUUGAGAAAAUCACAAAUGCG .....(((..((((((((-(......(((..((((((((.....))))))))..)))(((((((....)))))))..))).)))))).)))............. ( -35.10, z-score = -2.97, R) >consensus ACCUGUUCGACAACUCAC_UUGCAAAUGAUCUUAGCU___GGACCCAGCUGAUCUCAGAUCUGCGAUCGCAGAUCUCAGUCGAGUUGAGAAAAUCACAAAUGCG .....................(((..(((((((((((...(....).(((((.....(((((((....))))))))))))..))))))))...)))....))). (-19.96 = -20.28 + 0.32)
| Location | 12,469,440 – 12,469,530 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Shannon entropy | 0.24262 |
| G+C content | 0.42364 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.46 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12469440 90 + 23011544 GAUCGCAGAUCUGAGAUCAGCUGGCUCC---AGCUAAGAUCAUUUGCAAAGUGAGUUGUCGAACAGGUCCCCGAAAUAAAUGAAAAAAAAAAA ..(((..((((((.(((((((((....)---))))..)))).(((((((......))).))))))))))..)))................... ( -23.80, z-score = -2.76, R) >droEre2.scaffold_4929 13691168 79 - 26641161 GAUUUGAGAUUUGAGAUCAGCUGGGUCC---AGCUAAGAUCAUUUGCAA-GUAAGUGGUCGAACAGGUCCCCGAAAUAAAUAA---------- ..((((.((((((.....(((((....)---))))..(((((((((...-.)))))))))...))))))..))))........---------- ( -23.10, z-score = -2.81, R) >droYak2.chr2L 8906554 77 + 22324452 GAUCGCAGAUUUGAGAUCAGCUGUGUCC---AGCUAAGAUCAUUUGCAA-GUGAGUUGUCGAACAGGUCCCCGAAAUAAAU------------ ..(((..((((((.(((((((((....)---))))..)))).(((((((-.....))).))))))))))..))).......------------ ( -19.10, z-score = -1.71, R) >droSec1.super_16 660290 92 + 1878335 GAUCGCAGAUCUGAGAUCAGCUGGGUCCACCAGCUAAGAUCAUUUGCAA-GUGAGUUGUCGAACAGGUCCCCGAAAUAAAUGAAAAAUAAAUA ..(((..((((((.((((((((((.....))))))..)))).(((((((-.....))).))))))))))..)))................... ( -26.30, z-score = -3.07, R) >droSim1.chr2L 12278442 92 + 22036055 GAUCGCAGAUCUGAGAUCAGCUGGGUCCACCAGCUAAGAUCAUUUGCAA-GUGAGUUGUCGAACAGGUCCCCGAAAUAAAUGAAAAAUAAAUA ..(((..((((((.((((((((((.....))))))..)))).(((((((-.....))).))))))))))..)))................... ( -26.30, z-score = -3.07, R) >consensus GAUCGCAGAUCUGAGAUCAGCUGGGUCC___AGCUAAGAUCAUUUGCAA_GUGAGUUGUCGAACAGGUCCCCGAAAUAAAUGAAAAA_AAA_A ..(((..((((((.((((((((.........))))..)))).(((((((......))).))))))))))..)))................... (-15.46 = -15.46 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:18 2011