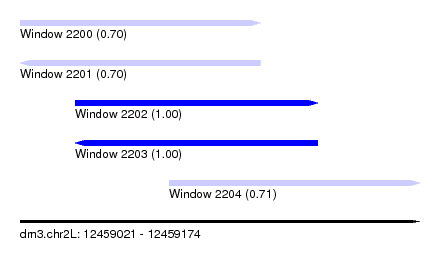
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,459,021 – 12,459,174 |
| Length | 153 |
| Max. P | 0.999542 |

| Location | 12,459,021 – 12,459,113 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Shannon entropy | 0.30722 |
| G+C content | 0.40614 |
| Mean single sequence MFE | -18.80 |
| Consensus MFE | -10.35 |
| Energy contribution | -11.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
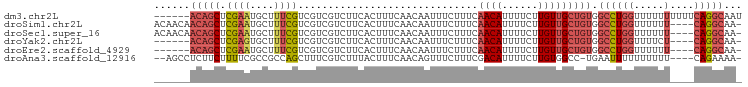
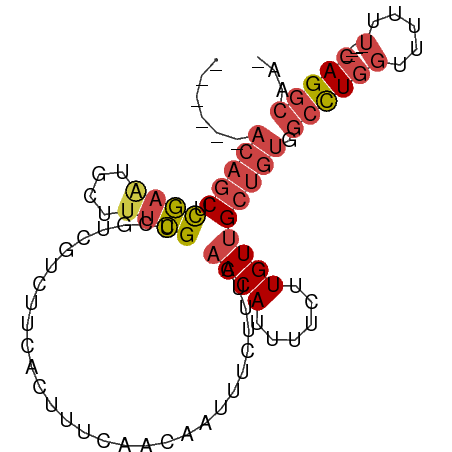
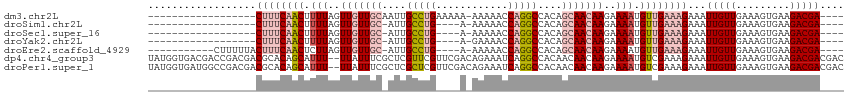
>dm3.chr2L 12459021 92 + 23011544 ------ACAGCUCGAAUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUUUUUUUCAGGCAAU ------(((((.((((....))))..............................((((......))))))))).((((((.........))))))... ( -18.70, z-score = -2.46, R) >droSim1.chr2L 12266939 93 + 22036055 ACAACAACAGCUCGAAUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUUU----CAGGCAA- ......(((((.((((....))))..............................((((......))))))))).((((((.....)----)))))..- ( -19.80, z-score = -2.25, R) >droSec1.super_16 650051 93 + 1878335 ACAACAACAGCUCGAAUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUUU----CAGGCAA- ......(((((.((((....))))..............................((((......))))))))).((((((.....)----)))))..- ( -19.80, z-score = -2.25, R) >droYak2.chr2L 8896096 87 + 22324452 ------ACAGCUCGAGUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUCU----CAGGCAA- ------(((((..(((((....((....))....)))))...............((((......))))))))).((((((.....)----)))))..- ( -21.10, z-score = -2.80, R) >droEre2.scaffold_4929 13681134 87 - 26641161 ------ACAGCUCGAAUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUUU----CAGGCAA- ------(((((.((((....))))..............................((((......))))))))).((((((.....)----)))))..- ( -19.50, z-score = -2.55, R) >droAna3.scaffold_12916 3248731 90 - 16180835 --AGCCUCUUCUUUUCGCCGCCAGCUUUCGUCUUUACUUUCAACAGUUUCUUUCGACAUUUUCUUGUGGCC-UGAAUUUUUUUUUU----CAGAAAA- --..............(((((.((...(((.....(((......)))......)))......)).)))))(-((((........))----)))....- ( -13.90, z-score = -1.21, R) >consensus ______ACAGCUCGAAUGCUUUCGUCGUCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUUU____CAGGCAA_ ......(((((.((((....))))..............................((((......))))))))).(((((...........)))))... (-10.35 = -11.35 + 1.00)
| Location | 12,459,021 – 12,459,113 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Shannon entropy | 0.30722 |
| G+C content | 0.40614 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -11.00 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12459021 92 - 23011544 AUUGCCUGAAAAAAAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCAUUCGAGCUGU------ ...(((((...........))))).(((((((((......))))(((.(...((((((...((....))..))))))...).)))..)))))------ ( -20.10, z-score = -2.68, R) >droSim1.chr2L 12266939 93 - 22036055 -UUGCCUG----AAAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCAUUCGAGCUGUUGUUGU -..(((((----.......)))))(((..((((((.......((....))..))))))...)))...((((((((...(((......))))))))))) ( -24.20, z-score = -2.94, R) >droSec1.super_16 650051 93 - 1878335 -UUGCCUG----AAAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCAUUCGAGCUGUUGUUGU -..(((((----.......)))))(((..((((((.......((....))..))))))...)))...((((((((...(((......))))))))))) ( -24.20, z-score = -2.94, R) >droYak2.chr2L 8896096 87 - 22324452 -UUGCCUG----AGAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCACUCGAGCUGU------ -..(((((----.......))))).(((((((((......))))........((((((...((....))..))))))..........)))))------ ( -19.10, z-score = -1.92, R) >droEre2.scaffold_4929 13681134 87 + 26641161 -UUGCCUG----AAAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCAUUCGAGCUGU------ -..(((((----.......))))).(((((((((......))))(((.(...((((((...((....))..))))))...).)))..)))))------ ( -20.80, z-score = -2.84, R) >droAna3.scaffold_12916 3248731 90 + 16180835 -UUUUCUG----AAAAAAAAAAUUCA-GGCCACAAGAAAAUGUCGAAAGAAACUGUUGAAAGUAAAGACGAAAGCUGGCGGCGAAAAGAAGAGGCU-- -....(((----((........))))-)(((...........((....))...(((((..(((..........)))..))))).........))).-- ( -16.40, z-score = -0.65, R) >consensus _UUGCCUG____AAAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGACGACGAAAGCAUUCGAGCUGU______ ...(((((...........))))).(((((((((......))))........((((((...((....))..))))))..........)))))...... (-11.00 = -12.28 + 1.28)

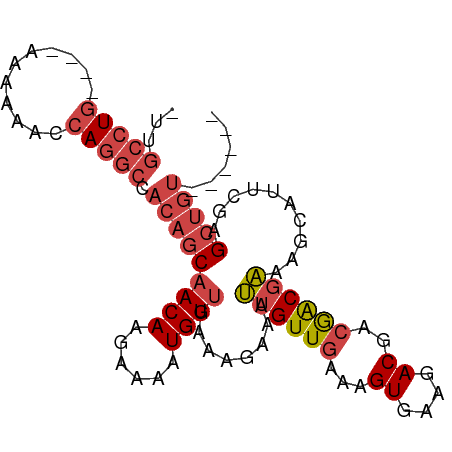
| Location | 12,459,042 – 12,459,135 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.40162 |
| G+C content | 0.38432 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -18.36 |
| Energy contribution | -19.09 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
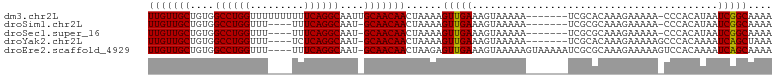
>dm3.chr2L 12459042 93 + 23011544 ----UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUU-UUUUUCAGGCAAUUGCAACAACUAAAAGUUGAAAG------------------ ----......................((((((((.(((..(((((((....((((((.....-....))))))....)))))))..))).))))))))------------------ ( -28.90, z-score = -5.69, R) >droSim1.chr2L 12266966 88 + 22036055 ----UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUU-U----CAGGCAAU-GCAACAACUAAAAGUUGAAAG------------------ ----......................((((((((.(((..(((((((....((((((.....-)----)))))...-)))))))..))).))))))))------------------ ( -29.30, z-score = -5.93, R) >droSec1.super_16 650078 88 + 1878335 ----UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUU-U----CAGGCAAU-GCAACAACUAAAAGUUGAAAG------------------ ----......................((((((((.(((..(((((((....((((((.....-)----)))))...-)))))))..))).))))))))------------------ ( -29.30, z-score = -5.93, R) >droYak2.chr2L 8896117 88 + 22324452 ----UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUC-U----CAGGCAAU-GCAACAACUAAAAGUUGAAAG------------------ ----......................((((((((.(((..(((((((....((((((.....-)----)))))...-)))))))..))).))))))))------------------ ( -28.50, z-score = -5.80, R) >droEre2.scaffold_4929 13681155 95 - 26641161 ----UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUU-U----CAGGCAAU-GCAACAACUAAGAGUUGAAAGUAAAAAG----------- ----...................(((((((((((.(((..(((((((....((((((.....-)----)))))...-)))))))..))).)))))))).)))...----------- ( -29.90, z-score = -4.81, R) >dp4.chr4_group3 3149098 114 + 11692001 GUCGUCGUCUUCACUUUCAACAAUUUCUUUCGACAUUUUCUUGUUGUUGUGGCCUGAUUUCUGUCGAACGAACGAGCGAAAUAA--AAAUGCUGUGCGUCGUCGGUCGUCACCAUA ..............................(((((......))))).(((((..((((..(((.(((.((.((.((((......--...)))))).))))).)))..))))))))) ( -21.90, z-score = 1.15, R) >droPer1.super_1 4631804 114 + 10282868 GUCGUCGUCUUCACUUUCAACAAUUUCUUUCGACAUUUUCUUGUUGUUGUGGCCUGAUUUCUGUCGAACGAGCGAGCGAAAUAA--AAAUGCUGUGCGUCGUCGGCCAUCACCAUA ...((((.......................)))).......(((.((.((((((.(((....)))((.((((((((((......--...)))).))).))))))))))).)).))) ( -24.20, z-score = 0.41, R) >consensus ____UCGUCUUCACUUUCAACAAUUUCUUUCAACAUUUUCUUGUUGCUGUGGCCUGGUUUUU_U____CAGGCAAU_GCAACAACUAAAAGUUGAAAG__________________ ..........................((((((((.(((..(((((((.((.(((((............))))).)).)))))))..))).)))))))).................. (-18.36 = -19.09 + 0.72)

| Location | 12,459,042 – 12,459,135 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Shannon entropy | 0.40162 |
| G+C content | 0.38432 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -16.48 |
| Energy contribution | -18.39 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12459042 93 - 23011544 ------------------CUUUCAACUUUUAGUUGUUGCAAUUGCCUGAAAAA-AAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA---- ------------------((((((((((((..(((((((....(((((.....-......)))))....))))))).)))).))))))))...(((((.........)))))---- ( -26.80, z-score = -4.22, R) >droSim1.chr2L 12266966 88 - 22036055 ------------------CUUUCAACUUUUAGUUGUUGC-AUUGCCUG----A-AAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA---- ------------------((((((((((((..(((((((-...(((((----.-......)))))....))))))).)))).))))))))...(((((.........)))))---- ( -27.10, z-score = -4.29, R) >droSec1.super_16 650078 88 - 1878335 ------------------CUUUCAACUUUUAGUUGUUGC-AUUGCCUG----A-AAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA---- ------------------((((((((((((..(((((((-...(((((----.-......)))))....))))))).)))).))))))))...(((((.........)))))---- ( -27.10, z-score = -4.29, R) >droYak2.chr2L 8896117 88 - 22324452 ------------------CUUUCAACUUUUAGUUGUUGC-AUUGCCUG----A-GAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA---- ------------------((((((((((((..(((((((-...(((((----.-......)))))....))))))).)))).))))))))...(((((.........)))))---- ( -27.10, z-score = -4.19, R) >droEre2.scaffold_4929 13681155 95 + 26641161 -----------CUUUUUACUUUCAACUCUUAGUUGUUGC-AUUGCCUG----A-AAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA---- -----------.((((((((((((((((((.(((((((.-...(((((----.-......)))))..))))))))))).(((.((....)).)))))))))))))))))...---- ( -32.80, z-score = -5.42, R) >dp4.chr4_group3 3149098 114 - 11692001 UAUGGUGACGACCGACGACGCACAGCAUUU--UUAUUUCGCUCGUUCGUUCGACAGAAAUCAGGCCACAACAACAAGAAAAUGUCGAAAGAAAUUGUUGAAAGUGAAGACGACGAC ....(((.((........))))).......--.....(((.(((((.((..((......))..))(((..((((((.......((....))..))))))...)))..)))))))). ( -21.10, z-score = 1.28, R) >droPer1.super_1 4631804 114 - 10282868 UAUGGUGAUGGCCGACGACGCACAGCAUUU--UUAUUUCGCUCGCUCGUUCGACAGAAAUCAGGCCACAACAACAAGAAAAUGUCGAAAGAAAUUGUUGAAAGUGAAGACGACGAC .....((.((((((((((.((..(((....--.......))).))))))).((......)).))))))).((((((.......((....))..))))))................. ( -24.50, z-score = 0.32, R) >consensus __________________CUUUCAACUUUUAGUUGUUGC_AUUGCCUG____A_AAAAACCAGGCCACAGCAACAAGAAAAUGUUGAAAGAAAUUGUUGAAAGUGAAGACGA____ ..................((((((((.(((..(((((((....(((((............)))))....)))))))..))).))))))))...(((((.........))))).... (-16.48 = -18.39 + 1.90)
| Location | 12,459,078 – 12,459,174 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Shannon entropy | 0.14841 |
| G+C content | 0.37349 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -20.07 |
| Energy contribution | -19.83 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
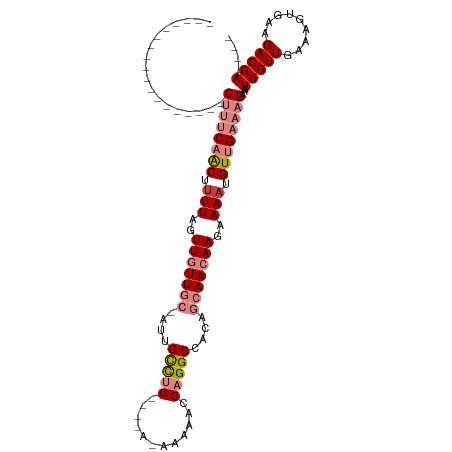
>dm3.chr2L 12459078 96 + 23011544 UUGUUGCUGUGGCCUGGUUUUUUUUUUCAGGCAAUUGCAACAACUAAAAGUUGAAAGUAAAAA-------UCGCACAAAGAAAAA-CCCACAUAAUCGGCAAAA ((((((.((((....(((((((((((....((..((((..((((.....))))...))))...-------..))..)))))))))-))..))))..)))))).. ( -19.90, z-score = -0.46, R) >droSim1.chr2L 12267002 91 + 22036055 UUGUUGCUGUGGCCUGGUUU----UUUCAGGCAAU-GCAACAACUAAAAGUUGAAAGUAAAAA-------UCGCGCAAAGAAAAA-CCCACAUAAUCGGCAAAA ((((((.((((((((((...----..))))))..(-((..((((.....))))...((.....-------..)))))........-..))))....)))))).. ( -21.90, z-score = -1.29, R) >droSec1.super_16 650114 91 + 1878335 UUGUUGCUGUGGCCUGGUUU----UUUCAGGCAAU-GCAACAACUAAAAGUUGAAAGUAAAAA-------UCGCGCAAAGAAAAA-CCCACAUAAUCGGCAAAA ((((((.((((((((((...----..))))))..(-((..((((.....))))...((.....-------..)))))........-..))))....)))))).. ( -21.90, z-score = -1.29, R) >droYak2.chr2L 8896153 92 + 22324452 UUGUUGCUGUGGCCUGGUUU----UCUCAGGCAAU-GCAACAACUAAAAGUUGAAAGUAAAAA-------UCGCACAAAGAAAAAGCCCACAAAAUCAGCUAAA (((((((....((((((...----..))))))...-))))))).....((((((.........-------..((...........))........))))))... ( -20.91, z-score = -1.32, R) >droEre2.scaffold_4929 13681191 99 - 26641161 UUGUUGCUGUGGCCUGGUUU----UUUCAGGCAAU-GCAACAACUAAGAGUUGAAAGUAAAAAGUAAAAAUCGCGCAAAGAAAAAGUCCACAAAAUCAGCAAAA ((((((.((((((((((...----..))))))..(-((..((((.....))))..........((.......)))))...........))))....)))))).. ( -22.70, z-score = -1.56, R) >consensus UUGUUGCUGUGGCCUGGUUU____UUUCAGGCAAU_GCAACAACUAAAAGUUGAAAGUAAAAA_______UCGCGCAAAGAAAAA_CCCACAUAAUCGGCAAAA (((((((.((.((((((.........)))))).)).)))))))......(((((................((.......))..............))))).... (-20.07 = -19.83 + -0.24)
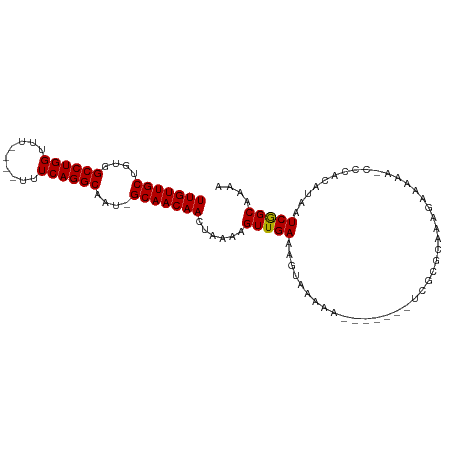
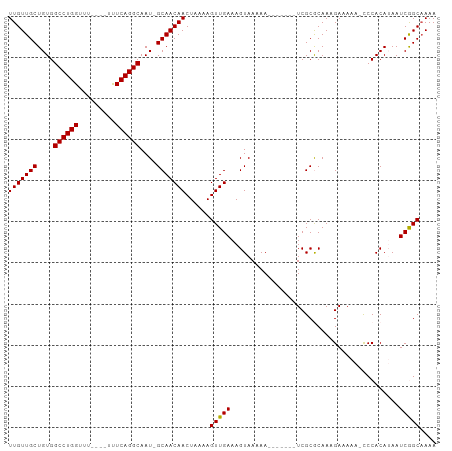
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:17 2011