
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,391,510 – 12,391,604 |
| Length | 94 |
| Max. P | 0.550715 |

| Location | 12,391,510 – 12,391,604 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.67559 |
| G+C content | 0.46573 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -8.85 |
| Energy contribution | -8.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 12391510 94 - 23011544 ----GUCGACUACUUGCUGACCCCUUCGACCGACGUCUAGCGCCUGGAU----CUGGCAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----((((.(.....).)))).....((((.(((((.....(((.....----..))).........-----.((((((((.....))))))))..)))))))))..--- ( -26.40, z-score = -1.72, R) >droSim1.chr2L 12197106 94 - 22036055 ----GUCGAGUCCUUGUUGACCCCUUCGACCAACGUCUAGCGCCUGGAU----CUGGCAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----(((((.......))))).....((((..((((.....(((.....----..))).........-----.((((((((.....))))))))..)))).))))..--- ( -22.70, z-score = -0.81, R) >droSec1.super_16 582439 94 - 1878335 ----GUCGAGUCCUUGUUGACCCCUUCGACCAACGUCUAGCGCCUGGAU----CUGGCAUUUAAAAA-----CGUUAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----(((((.......))))).....((((..((((.....(((.....----..))).........-----.((((((((.....))))))))..)))).))))..--- ( -20.60, z-score = -0.19, R) >droYak2.chr2L 8827290 94 - 22324452 ----GUCGAGUCCUUUGUGACCCCUUCGACCAACGUCUAGCUCCUGGAU----CUGGCAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----............(((((..(...(.(((..((((((...))))))----.)))).........-----.((((((((.....))))))))....)..))))).--- ( -21.90, z-score = -1.04, R) >droEre2.scaffold_4929 13616223 94 + 26641161 ----GUCGAGUCCUUUCUGACCCCUUCGACCAGCGUCUAGCUCCUGGAU----GUGGCAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----((((((((......)))....))))).((((.(..((..(.....----)..)).........-----.((((((((.....)))))))).......).))))--- ( -23.70, z-score = -1.29, R) >droAna3.scaffold_12916 3174856 92 + 16180835 --GGGUCGGACACCUCCCUCUCUUCUCAGCCGAUUUCU----UCUGGAU----GUGGCAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- --(((..(((....)))..)))......((((..(((.----...))).----.)))).........-----.((((((((.....)))))))).............--- ( -21.20, z-score = -0.71, R) >droPer1.super_1 3087688 98 + 10282868 ----GUUGGGUCUUCUUCUCCAGCUUCUGCUGCUCCUCUGUGUAUGGAUGGGGCUGACAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----((((((........))))))....((.((((((((......))).))))).............-----.((((((((.....))))))))..........)).--- ( -26.00, z-score = -1.53, R) >dp4.chr4_group3 5988368 98 + 11692001 ----GUUGGGUCUUCUUCUCCAGCUUCUGCUGCUCCUCUGUGUAUGGAUGGGGCUGACAUUUAAAAA-----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU--- ----((((((........))))))....((.((((((((......))).))))).............-----.((((((((.....))))))))..........)).--- ( -26.00, z-score = -1.53, R) >droWil1.scaffold_180708 10127272 102 - 12563649 GCUGGUUGUUGUCCUUUAGUGUCCUGAACAUUGCGCCCCUCGUACUUUU---GCUGACAUUUAAAAA-----UGUCAUUUGUUAUUCAAGUGGCAUACGUUGUGUCGCUU ...((.(((.....(((((....)))))....))).))...........---((.(((((......(-----(((((((((.....)))))))))).....))))))).. ( -21.70, z-score = -0.84, R) >droVir3.scaffold_12963 17506401 80 - 20206255 --------------UUGAG-UCAAAGUCU--GCGUCUCGGAGUCCGGUU---GCUGACAUUUAAAAAA----CGUCAUUUGUUAUUCAAGUGGCAUACGUCGCU------ --------------.....-.....(((.--(((.(.(((...)))).)---)).)))..........----.((((((((.....))))))))..........------ ( -18.30, z-score = -0.61, R) >droMoj3.scaffold_6500 22548715 84 - 32352404 -----UAGA---GUCCAAG-CUUCAGUCUC---GGCUGACAGUCC-GUU---GCUGACAUUUAAAAAAA----GUCAUUUGUUAUUCAAGUGGCAUACGUCGCU------ -----....---.....((-(.(((((..(---(((.....).))-)..---)))))............----((((((((.....)))))))).......)))------ ( -21.00, z-score = -1.73, R) >droGri2.scaffold_15252 14938619 96 - 17193109 ----CUGGACUCGUCCAAGUCUGAAGUCUCUGUCGCUCGGAGUCC-GUU---GCUGACAUUUAAAAAAAUAACGUCAUUUGUUAUUCAAGUGGCAUACGUCGCU------ ----.((((....))))........(.(((((.....))))).).-...---((.(((((((....))))...((((((((.....))))))))....))))).------ ( -22.20, z-score = -0.90, R) >consensus ____GUCGAGUCCUUUUUGACCCCUUCGACCGACGCCUAGCGUCUGGAU____CUGACAUUUAAAAA_____CGUCAUUUGUUAUUCAAGUGGCAUACGUCGUCGCU___ .........................................................................((((((((.....))))))))................ ( -8.85 = -8.78 + -0.08)
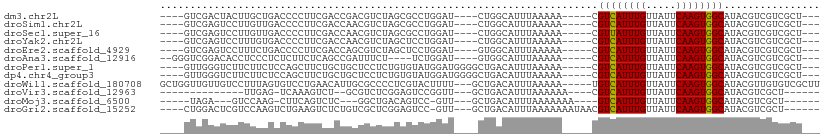

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:11 2011