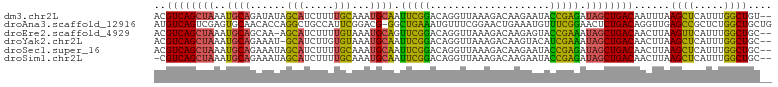
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,378,265 – 12,378,366 |
| Length | 101 |
| Max. P | 0.976675 |

| Location | 12,378,265 – 12,378,366 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Shannon entropy | 0.45244 |
| G+C content | 0.42522 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
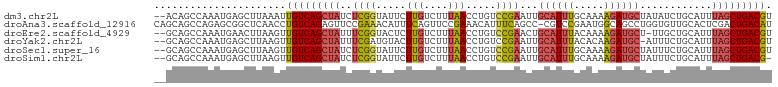
>dm3.chr2L 12378265 101 + 23011544 --ACAGCCAAAUGAGCUUAAAUUGUCAGCUAUCUCGGUAUUCUUGUCUUUAACCUGUCCGAAUUGCAUUUGCAAAAGAUGCUAUAUCUGCAUUUAGCUGACGU --..((((....).)))......((((((((..((((.....(((....))).....)))).((((....))))..(((((.......))))))))))))).. ( -24.20, z-score = -1.78, R) >droAna3.scaffold_12916 3161954 102 - 16180835 CAGCAGCCAGAGCGGCUCAACCUGUCAGAGUUCCGAAACAUUUCAGUUCCGAAACAUUUCAGCC-CGUCCGAAUGGCAGCCUGGUGUUGCACUCGACUGACAU ..(.((((.....)))))....((((((.(((.((.(((......))).)).))).........-....(((...(((((.....)))))..))).)))))). ( -25.80, z-score = 0.12, R) >droEre2.scaffold_4929 13603242 100 - 26641161 --GCAGCCAAAUGAACUUAAGUUGUCAGCUAUUUCGGUACUCUUGUCUUUAACCUGUCCGAACUGCAUUUACAAAAGAUGCU-UUGCUGCAUUUAGCUGACGU --.((((.((((((((....)))..((((...(((((.((..(((....)))...)))))))..((((((.....)))))).-..))))))))).)))).... ( -23.70, z-score = -1.28, R) >droYak2.chr2L 8813742 100 + 22324452 --GCAGCCAAAUGAGCUUAAGUUGUCAGCUAUUUCGAUGUACUUGUCUUUAACCUGUCCGAAUUGCAUUUACACAAGAUGC-AUUUCUGCAUUUAGCUGACGU --..((((....).)))......((((((((....((((((...(.(........).).(((.(((((((.....))))))-).))))))))))))))))).. ( -24.30, z-score = -1.43, R) >droSec1.super_16 569300 101 + 1878335 --GCAGCCAAAUGAGCUUAAGUUGUCAGCUAUCUCGGUAUUCUUGUCUUUAACCUGUCCGAAUUGCAUUUGCAAAAGAUGCUAUUUCUGCAUUUAGCUGACGU --..((((....).)))......((((((((..((((.....(((....))).....)))).((((....))))..(((((.......))))))))))))).. ( -24.20, z-score = -1.06, R) >droSim1.chr2L 12181685 100 + 22036055 --GCAGCCAAAUGAGCUUAAGUUGUCAGCUAUCUCGGUAUUCUUGUCUUUAACCUGUCCGAAUUGCAUUUGCAAAAGAUGCUAUUUCUGCAUUUAGCUGACG- --..((((....).)))......((((((((..((((.....(((....))).....)))).((((....))))..(((((.......))))))))))))).- ( -24.20, z-score = -1.30, R) >consensus __GCAGCCAAAUGAGCUUAAGUUGUCAGCUAUCUCGGUAUUCUUGUCUUUAACCUGUCCGAAUUGCAUUUGCAAAAGAUGCUAUUUCUGCAUUUAGCUGACGU ......................(((((((((..((((.....(((....))).....))))...((((((.....))))))............))))))))). (-16.32 = -16.52 + 0.20)
| Location | 12,378,265 – 12,378,366 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Shannon entropy | 0.45244 |
| G+C content | 0.42522 |
| Mean single sequence MFE | -27.55 |
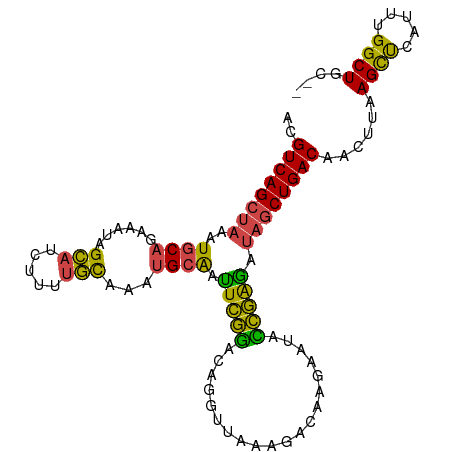
| Consensus MFE | -21.46 |
| Energy contribution | -20.13 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
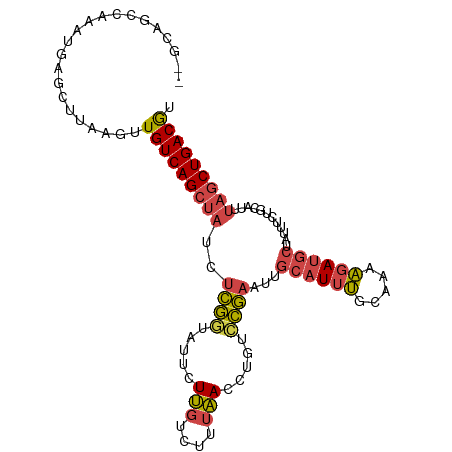
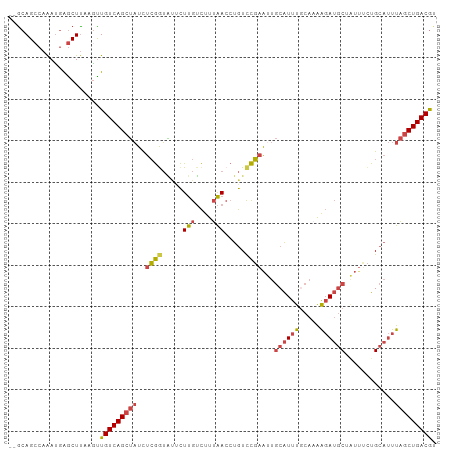
>dm3.chr2L 12378265 101 - 23011544 ACGUCAGCUAAAUGCAGAUAUAGCAUCUUUUGCAAAUGCAAUUCGGACAGGUUAAAGACAAGAAUACCGAGAUAGCUGACAAUUUAAGCUCAUUUGGCUGU-- ..((((((((..((((......(((.....)))...)))).(((((....(((...))).......))))).))))))))......((((.....))))..-- ( -25.40, z-score = -1.76, R) >droAna3.scaffold_12916 3161954 102 + 16180835 AUGUCAGUCGAGUGCAACACCAGGCUGCCAUUCGGACG-GGCUGAAAUGUUUCGGAACUGAAAUGUUUCGGAACUCUGACAGGUUGAGCCGCUCUGGCUGCUG ..(.((((((((((......).(((((((..(((((.(-..(((((((((((((....)))))))))))))..))))))..)))..)))))))).)))))).. ( -37.70, z-score = -1.93, R) >droEre2.scaffold_4929 13603242 100 + 26641161 ACGUCAGCUAAAUGCAGCAA-AGCAUCUUUUGUAAAUGCAGUUCGGACAGGUUAAAGACAAGAGUACCGAAAUAGCUGACAACUUAAGUUCAUUUGGCUGC-- ....((((((((((((((..-.((((.........))))..(((((((..(((...)))....)).)))))...))))..(((....))))))))))))).-- ( -25.40, z-score = -1.23, R) >droYak2.chr2L 8813742 100 - 22324452 ACGUCAGCUAAAUGCAGAAAU-GCAUCUUGUGUAAAUGCAAUUCGGACAGGUUAAAGACAAGUACAUCGAAAUAGCUGACAACUUAAGCUCAUUUGGCUGC-- ..((((((((.(((((....)-)))).((((......))))(((((((..(((...)))..))...))))).))))))))......((((.....))))..-- ( -25.60, z-score = -1.53, R) >droSec1.super_16 569300 101 - 1878335 ACGUCAGCUAAAUGCAGAAAUAGCAUCUUUUGCAAAUGCAAUUCGGACAGGUUAAAGACAAGAAUACCGAGAUAGCUGACAACUUAAGCUCAUUUGGCUGC-- ..((((((((..(((((((........))))))).......(((((....(((...))).......))))).))))))))......((((.....))))..-- ( -25.60, z-score = -1.75, R) >droSim1.chr2L 12181685 100 - 22036055 -CGUCAGCUAAAUGCAGAAAUAGCAUCUUUUGCAAAUGCAAUUCGGACAGGUUAAAGACAAGAAUACCGAGAUAGCUGACAACUUAAGCUCAUUUGGCUGC-- -.((((((((..(((((((........))))))).......(((((....(((...))).......))))).))))))))......((((.....))))..-- ( -25.60, z-score = -1.75, R) >consensus ACGUCAGCUAAAUGCAGAAAUAGCAUCUUUUGCAAAUGCAAUUCGGACAGGUUAAAGACAAGAAUACCGAGAUAGCUGACAACUUAAGCUCAUUUGGCUGC__ ..((((((((..((((......(((.....)))...)))).(((((....................))))).))))))))......((((.....)))).... (-21.46 = -20.13 + -1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:08 2011