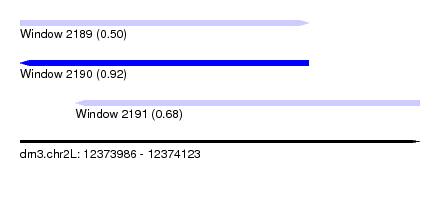
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,373,986 – 12,374,123 |
| Length | 137 |
| Max. P | 0.921710 |

| Location | 12,373,986 – 12,374,085 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.44205 |
| G+C content | 0.57747 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -13.15 |
| Energy contribution | -12.79 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
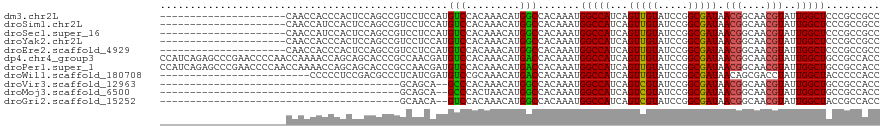
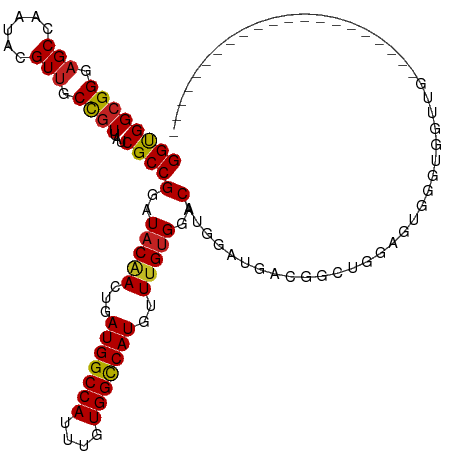
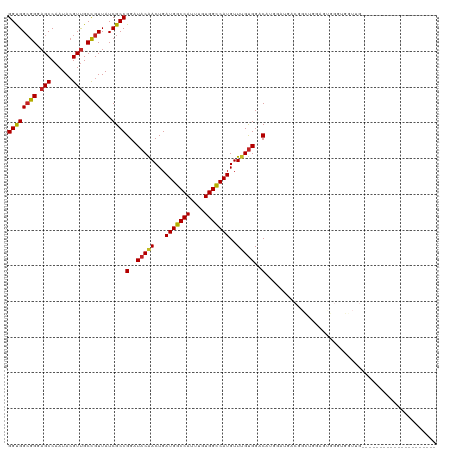
>dm3.chr2L 12373986 99 + 23011544 ---------------------CAACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCC ---------------------...............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))........(((....)))... ( -27.00, z-score = -1.53, R) >droSim1.chr2L 12177352 99 + 22036055 ---------------------CAACCAUCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCC ---------------------...............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))........(((....)))... ( -27.00, z-score = -1.65, R) >droSec1.super_16 565014 99 + 1878335 ---------------------CAACCAUCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCC ---------------------...............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))........(((....)))... ( -27.00, z-score = -1.65, R) >droYak2.chr2L 8809458 99 + 22324452 ---------------------CAACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCC ---------------------...............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))........(((....)))... ( -27.00, z-score = -1.53, R) >droEre2.scaffold_4929 13598891 99 - 26641161 ---------------------CAACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCC ---------------------...............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))........(((....)))... ( -27.00, z-score = -1.53, R) >dp4.chr4_group3 5968277 120 - 11692001 CCAUCAGAGCCCGAACCCCAACCAAAACCAGCAGCACCCGCCAACGAUGUCCACAAACAUGACCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACC ..............................(((((..(((.((((((((.(((................))).)))).))))....)))(((((.((....))))))))))))....... ( -26.39, z-score = -0.52, R) >droPer1.super_1 3067872 120 - 10282868 CCAUCAGAGCCCGAACCCCAACCAAAACCAGCAGCACCCGCCAACGAUGUCCACAAACAUGACCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACC ..............................(((((..(((.((((((((.(((................))).)))).))))....)))(((((.((....))))))))))))....... ( -26.39, z-score = -0.52, R) >droWil1.scaffold_180708 10106489 95 + 12563649 -------------------------CCCCCUCCGACGCCCUCAUCGAUGUCCGCAAACAUGACCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACAGCGACCUAUUGGCUACCCCCACC -------------------------...........(((......((((.(((................))).)))).((((((((....))).)))))........))).......... ( -14.99, z-score = 0.84, R) >droVir3.scaffold_12963 17478211 78 + 20206255 ----------------------------------------GCAGCA--GCCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACC ----------------------------------------((.(((--(((.......(((((((....)))))))..(((((.....))))).(((....)))...)))))).)).... ( -32.20, z-score = -3.62, R) >droMoj3.scaffold_6500 22524188 78 + 32352404 ----------------------------------------GCAGCA--GCCCACUAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACC ----------------------------------------((.(((--(((.......(((((((....)))))))..(((((.....))))).(((....)))...)))))).)).... ( -32.20, z-score = -3.55, R) >droGri2.scaffold_15252 14912655 78 + 17193109 ----------------------------------------GCAACA--GUCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUACCGCCACC ----------------------------------------......--(((.......(((((((....)))))))..((((....))))))).(((....)))..((((....)))).. ( -23.40, z-score = -1.92, R) >consensus _____________________CAACCACCCACUCCAGCCGUCAUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCACC ................................................(((.........))).......(((((...(((((.....))))).(((....)))..)))))......... (-13.15 = -12.79 + -0.36)
| Location | 12,373,986 – 12,374,085 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.44205 |
| G+C content | 0.57747 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
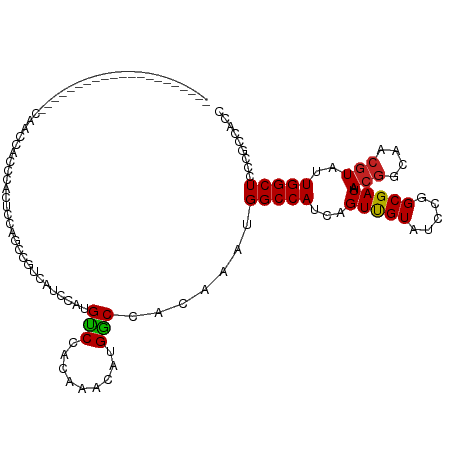
>dm3.chr2L 12373986 99 - 23011544 GGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGGUGGUUG--------------------- ((((((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))....))).--------------------- ( -36.40, z-score = -0.99, R) >droSim1.chr2L 12177352 99 - 22036055 GGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGAUGGUUG--------------------- ((((((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))....))).--------------------- ( -36.40, z-score = -1.34, R) >droSec1.super_16 565014 99 - 1878335 GGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGAUGGUUG--------------------- ((((((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))....))).--------------------- ( -36.40, z-score = -1.34, R) >droYak2.chr2L 8809458 99 - 22324452 GGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGGUGGUUG--------------------- ((((((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))....))).--------------------- ( -36.40, z-score = -0.99, R) >droEre2.scaffold_4929 13598891 99 + 26641161 GGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGGUGGUUG--------------------- ((((((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))....))).--------------------- ( -36.40, z-score = -0.99, R) >dp4.chr4_group3 5968277 120 + 11692001 GGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGUGGACAUCGUUGGCGGGUGCUGCUGGUUUUGGUUGGGGUUCGGGCUCUGAUGG ((((((((((((......)))))))))))).((((((.(((((.(((((((....)))))))....(..(.((((.......)))))..).......)))))..)))))).......... ( -45.80, z-score = -1.36, R) >droPer1.super_1 3067872 120 + 10282868 GGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGUGGACAUCGUUGGCGGGUGCUGCUGGUUUUGGUUGGGGUUCGGGCUCUGAUGG ((((((((((((......)))))))))))).((((((.(((((.(((((((....)))))))....(..(.((((.......)))))..).......)))))..)))))).......... ( -45.80, z-score = -1.36, R) >droWil1.scaffold_180708 10106489 95 - 12563649 GGUGGGGGUAGCCAAUAGGUCGCUGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGCGGACAUCGAUGAGGGCGUCGGAGGGGG------------------------- (((((.((..(((....)))..)).)))))((((.(.(((.....((((((....))))))))).).))).).((((((....))))))......------------------------- ( -29.80, z-score = -0.14, R) >droVir3.scaffold_12963 17478211 78 - 20206255 GGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGGC--UGCUGC---------------------------------------- ((((((((((((......))))))))))))(((...(((((...(((((((....)))))))..))))))))--......---------------------------------------- ( -35.40, z-score = -2.45, R) >droMoj3.scaffold_6500 22524188 78 - 32352404 GGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUAGUGGGC--UGCUGC---------------------------------------- ....(((((((((....(((..(((.......)))..)))(((((((((((....))))))..))))).)))--))))))---------------------------------------- ( -36.20, z-score = -2.69, R) >droGri2.scaffold_15252 14912655 78 - 17193109 GGUGGCGGUAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGAC--UGUUGC---------------------------------------- ((((((((((((......))))))))))))((.(..(((((...(((((((....)))))))..)))))..)--.))...---------------------------------------- ( -32.00, z-score = -2.49, R) >consensus GGUGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAUGACGGCUGGAGUGGGUGGUUG_____________________ ((((((((.(((......))).))))...))))(..(((((...(((((((....)))))))..)))))..)................................................ (-26.38 = -26.08 + -0.30)

| Location | 12,374,005 – 12,374,123 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Shannon entropy | 0.16701 |
| G+C content | 0.50941 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -33.97 |
| Energy contribution | -33.36 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
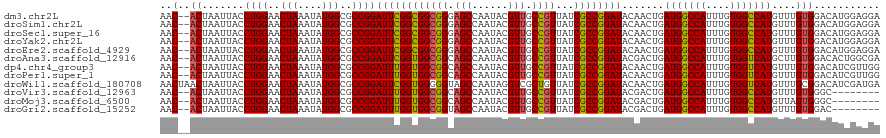
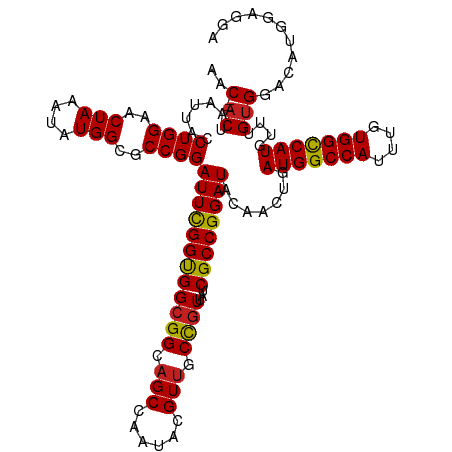
>dm3.chr2L 12374005 118 - 23011544 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ...--........(((((........(((((((((....))))(((....)))........))))).....))(..(((((...(((((((....)))))))..)))))..)....))). ( -40.82, z-score = -1.45, R) >droSim1.chr2L 12177371 118 - 22036055 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ...--........(((((........(((((((((....))))(((....)))........))))).....))(..(((((...(((((((....)))))))..)))))..)....))). ( -40.82, z-score = -1.45, R) >droSec1.super_16 565033 118 - 1878335 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ...--........(((((........(((((((((....))))(((....)))........))))).....))(..(((((...(((((((....)))))))..)))))..)....))). ( -40.82, z-score = -1.45, R) >droYak2.chr2L 8809477 118 - 22324452 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ...--........(((((........(((((((((....))))(((....)))........))))).....))(..(((((...(((((((....)))))))..)))))..)....))). ( -40.82, z-score = -1.45, R) >droEre2.scaffold_4929 13598910 118 + 26641161 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ...--........(((((........(((((((((....))))(((....)))........))))).....))(..(((((...(((((((....)))))))..)))))..)....))). ( -40.82, z-score = -1.45, R) >droAna3.scaffold_12916 3157813 118 + 16180835 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGUCAUGCUUGUGGACACUGGCGA ...--.........((((..(((....)))..))))....((((((((((((......))))))))))))(((((.(((((...(((((((....)))))))..)))))....))))).. ( -43.90, z-score = -2.28, R) >dp4.chr4_group3 5968317 118 + 11692001 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGUGGACAUCGUUGG ...--.(((((....((..((.(((((((((.((((....((((((((((((......)))))))))))).)))).(((((.((.....)).))))))))))))))))...))..))))) ( -39.70, z-score = -1.50, R) >droPer1.super_1 3067912 118 + 10282868 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGUGGACAUCGUUGG ...--.(((((....((..((.(((((((((.((((....((((((((((((......)))))))))))).)))).(((((.((.....)).))))))))))))))))...))..))))) ( -39.70, z-score = -1.50, R) >droWil1.scaffold_180708 10106504 120 - 12563649 AACUAACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGUGGGGGUAGCCAAUAGGUCGCUGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGCGGACAUCGAUGA .............((..((((......((...((((...((((((.((..(((....)))..)).))))))))))...))....(((((((....)))))))))))..)).......... ( -33.30, z-score = 0.20, R) >droVir3.scaffold_12963 17478217 110 - 20206255 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGGC-------- ...--........(((...((.(((((((((.((((....((((((((((((......)))))))))))).)))).(((((.((.....)).))))))))))))))))))).-------- ( -41.40, z-score = -2.16, R) >droMoj3.scaffold_6500 22524194 110 - 32352404 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUAGUGGGC-------- ..(--((((((...((((..(((....)))..))))((((((((((((((((......))))))))...)))))))).......(((((((....))))))))))))))...-------- ( -44.30, z-score = -3.00, R) >droGri2.scaffold_15252 14912661 110 - 17193109 AAC--ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUUGGUGGCGGUAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGAC-------- ...--........((..((((......((...((((....((((((((((((......)))))))))))).))))...))....(((((((....)))))))))))..))..-------- ( -39.00, z-score = -2.19, R) >consensus AAC__ACUAAUUACCUGGAACUAAAUAUGGCGCCGGAUUCGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGA ..............((((..(((....)))..))))((((((((((((.(((......))).))))...))))))))((((...(((((((....)))))))..))))............ (-33.97 = -33.36 + -0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:06 2011