| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,360,792 – 12,360,848 |
| Length | 56 |
| Max. P | 0.831785 |

| Location | 12,360,792 – 12,360,848 |
|---|---|
| Length | 56 |
| Sequences | 8 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 62.96 |
| Shannon entropy | 0.76773 |
| G+C content | 0.57357 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -9.15 |
| Energy contribution | -9.75 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.67 |
| Mean z-score | 0.03 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
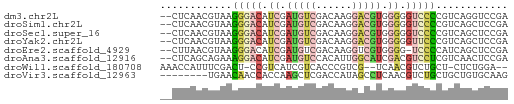
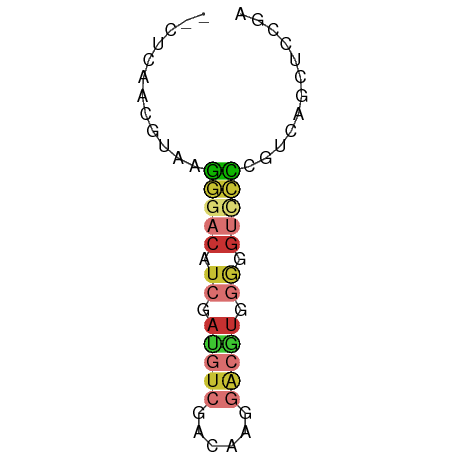
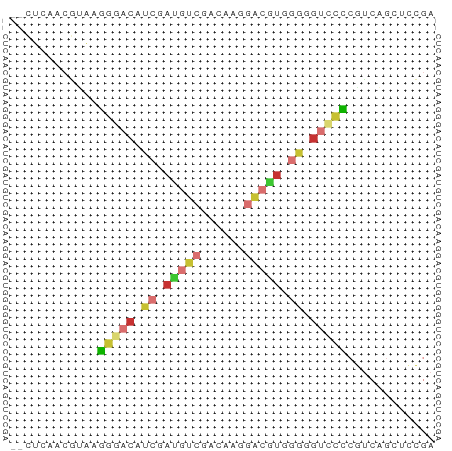
>dm3.chr2L 12360792 56 + 23011544 --CUCAACGUAAGGGACAUCGAUGUCGACAAGGACGUGGGGGUCCCCGUCAGGUCCGA --((..(((...(((((..(.(((((......))))).)..))))))))..))..... ( -18.60, z-score = -0.02, R) >droSim1.chr2L 12163732 56 + 22036055 --CUCAACGUAAGGGACAUCGAUGUCGACAAGGACGUGGGGGUCCCCGUCAGCUCCGA --....(((...(((((..(.(((((......))))).)..))))))))......... ( -17.70, z-score = 0.08, R) >droSec1.super_16 551659 56 + 1878335 --CUCAACGUAAGGGACAUCGAUGUCGACAAGGACGUGGGGGUCCCCGUCAGCUCCGA --....(((...(((((..(.(((((......))))).)..))))))))......... ( -17.70, z-score = 0.08, R) >droYak2.chr2L 8794303 56 + 22324452 --CUCAACGUAAGGGACAUCGAUGUCGACAAGGACGUGGGGGUUCCCGUCAGCUCCGA --....(((...(((((..(.(((((......))))).)..))))))))......... ( -14.80, z-score = 0.79, R) >droEre2.scaffold_4929 13585539 55 - 26641161 --CUUAACGUAAGGGACAUCGAUGUCGACAAGGUCGUGGGG-UCCCCAUCAGCUCCGA --..........(((((.((.(((.(......).))).)))-))))............ ( -12.70, z-score = 0.90, R) >droAna3.scaffold_12916 3144803 56 - 16180835 --CUCAGCAGAAAGGACAUCGAUGUCCACAUUGGCAUCGACGUCCUCGUCAACUCCGA --.......((.(((((.((((((((......)))))))).)))))..))........ ( -19.90, z-score = -3.08, R) >droWil1.scaffold_180708 10084548 52 + 12563649 AAACCAUUUCGACU-CCGUCAUCGUCACCCGUCG--UCAACGUCUGCU-CUCUGGA-- ...(((....(((.-..)))...(.((..(((..--...)))..))).-...))).-- ( -5.30, z-score = 1.06, R) >droVir3.scaffold_12963 17459517 50 + 20206255 --------UGAACAACCACCAAGCUCGACCAUAGCCUCAACGUCUGCUGCUGUGCAAG --------.....................((((((..((.....))..)))))).... ( -6.50, z-score = 0.47, R) >consensus __CUCAACGUAAGGGACAUCGAUGUCGACAAGGACGUGGGGGUCCCCGUCAGCUCCGA ............(((((.((.(((((......))))).)).)))))............ ( -9.15 = -9.75 + 0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:02 2011