| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,354,123 – 12,354,209 |
| Length | 86 |
| Max. P | 0.898398 |

| Location | 12,354,123 – 12,354,209 |
|---|---|
| Length | 86 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 76.38 |
| Shannon entropy | 0.38158 |
| G+C content | 0.37614 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -7.81 |
| Energy contribution | -9.50 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
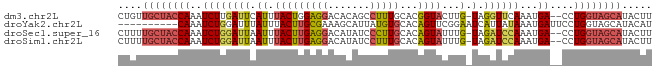
>dm3.chr2L 12354123 86 + 23011544 CUGUUGCUACCAAAUCUUGAUUCAUUUACUGGAGGACACAGCCUUUGCACGGUACUUG-UAGGUUCAAAUGA--CCUGGUAGCAUACUU ..(((((((((...........((..(((((((((......))))....)))))..))-.(((((.....))--)))))))))).)).. ( -22.60, z-score = -1.43, R) >droYak2.chr2L 8787670 79 + 22324452 ----------CAAAUCUGGAUUUAUUUACUUGCGAAAGCAUUAUGUGCACAGUUCGGAAUCAUUAUAAAUGAUUCCUGGUAGCAUACAU ----------....................(((....)))...(((((((.....((((((((.....))))))))..)).)))))... ( -19.50, z-score = -2.55, R) >droSec1.super_16 545040 86 + 1878335 CUUUUGCUACCAAAUCUGGAUUAAUUUACUUGAGGACAUAUCCCUUGCACAGUAUUUG-UAGAUCCAAAUGA--CCUGGUAGCAUACUU ....((((((((..((((((((.((.(((((((((.......)))))...))))...)-).))))))...))--..))))))))..... ( -23.70, z-score = -3.58, R) >droSim1.chr2L 12157108 86 + 22036055 CUUUUGCUACCAAAUCUGGAUUAAUUUACUUGAGGACAUAUCCUUUGCACAGUAUUUG-UAGAUCCAAAUGA--CCUGGUAGCAUACUU ....((((((((..((((((((.((.((((.(((((....))))).....))))...)-).))))))...))--..))))))))..... ( -24.80, z-score = -4.07, R) >consensus CUUUUGCUACCAAAUCUGGAUUAAUUUACUUGAGGACACAUCCUUUGCACAGUACUUG_UAGAUCCAAAUGA__CCUGGUAGCAUACUU ....((((((((..((((((((....(((((((((.......)))))...)))).......))))))...))....))))))))..... ( -7.81 = -9.50 + 1.69)

| Location | 12,354,123 – 12,354,209 |
|---|---|
| Length | 86 |
| Sequences | 4 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 76.38 |
| Shannon entropy | 0.38158 |
| G+C content | 0.37614 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -8.65 |
| Energy contribution | -10.53 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
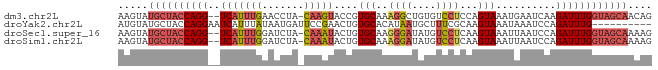
>dm3.chr2L 12354123 86 - 23011544 AAGUAUGCUACCAGG--UCAUUUGAACCUA-CAAGUACCGUGCAAAGGCUGUGUCCUCCAGUAAAUGAAUCAAGAUUUGGUAGCAACAG ..((.((((((((((--(((((((......-)))))...(..((...((((.(....)))))...))...)..)))))))))))))).. ( -24.50, z-score = -2.58, R) >droYak2.chr2L 8787670 79 - 22324452 AUGUAUGCUACCAGGAAUCAUUUAUAAUGAUUCCGAACUGUGCACAUAAUGCUUUCGCAAGUAAAUAAAUCCAGAUUUG---------- ((((..((.....((((((((.....)))))))).....))..))))..(((....)))....................---------- ( -16.10, z-score = -1.88, R) >droSec1.super_16 545040 86 - 1878335 AAGUAUGCUACCAGG--UCAUUUGGAUCUA-CAAAUACUGUGCAAGGGAUAUGUCCUCAAGUAAAUUAAUCCAGAUUUGGUAGCAAAAG .....((((((((((--((...(((((...-....((((.((...((((....)))))))))).....))))))))))))))))).... ( -26.00, z-score = -3.59, R) >droSim1.chr2L 12157108 86 - 22036055 AAGUAUGCUACCAGG--UCAUUUGGAUCUA-CAAAUACUGUGCAAAGGAUAUGUCCUCAAGUAAAUUAAUCCAGAUUUGGUAGCAAAAG .....((((((((((--((...(((((...-....((((.((...((((....)))))))))).....))))))))))))))))).... ( -26.00, z-score = -4.07, R) >consensus AAGUAUGCUACCAGG__UCAUUUGGAUCUA_CAAAUACUGUGCAAAGGAUAUGUCCUCAAGUAAAUUAAUCCAGAUUUGGUAGCAAAAG .....((((((((((..(((((((.......)))))....(((..((((....))))...)))..........)))))))))))).... ( -8.65 = -10.53 + 1.88)
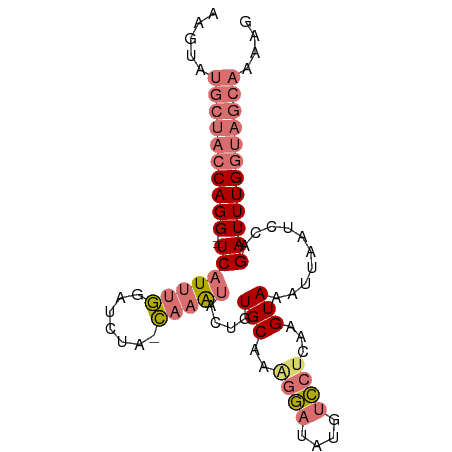
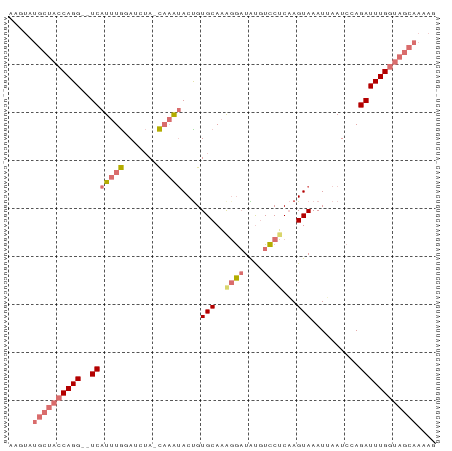
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:02 2011