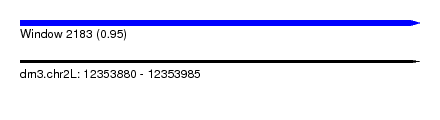
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 12,353,880 – 12,353,985 |
| Length | 105 |
| Max. P | 0.952237 |

| Location | 12,353,880 – 12,353,985 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
| Shannon entropy | 0.54521 |
| G+C content | 0.41462 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.36 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
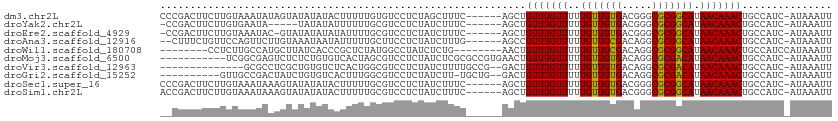
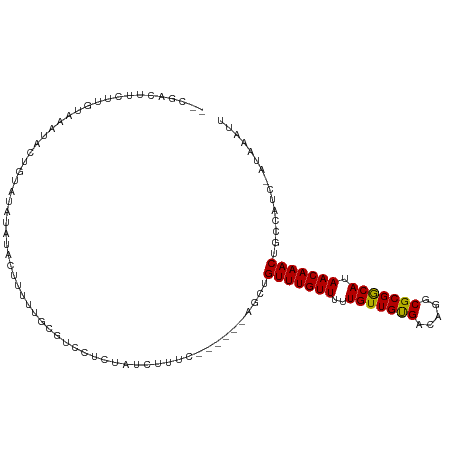
>dm3.chr2L 12353880 105 + 23011544 CCCGACUUCUUGUAAAUAUAGUAUAUAUACUUUUUGUGUCCUCUAGCUUUC------AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU ...(((.....(((.((((...)))).))).......)))...........------.((.(((((((..(((((((.....))))))).))))))).))....-....... ( -19.10, z-score = 0.01, R) >droYak2.chr2L 8787431 99 + 22324452 -CCGACUUCUUGUGAAUA-----UAUAUAUUUUUUGCGUCCUCUAUCUUUC------AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU -........((((((...-----............((((((....((...(------(((...........)))).)).))))))((((........)))).))-))))... ( -22.20, z-score = -1.71, R) >droEre2.scaffold_4929 13578830 103 - 26641161 -CCGACUUCUUGUAAAUAC-GUAUAUAUAUAUUUUGCGUCCUCUAUCUUUC------AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU -..((............((-(((...........)))))............------.((.(((((((..(((((((.....))))))).))))))).))..))-....... ( -19.10, z-score = -0.42, R) >droAna3.scaffold_12916 3136081 103 - 16180835 --CUUUCUGUUCCAGUUCUUGUAAAUAAUAUUUUUGCUUCCUCUAUCUUUG------AGCCGUUUGUUUUUGUUGCGACAGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU --...........((.....(((((.......))))).....)).....((------.((.(((((((..(((((((.....))))))).))))))).))))..-....... ( -21.40, z-score = -1.65, R) >droWil1.scaffold_180708 10076847 96 + 12563649 --------CCUCUUGCCAUGCUUAUCACCCGCUCUAUGGCCUAUCUCUG--------AACUGUUUGUUUUUGUUGCGACAGGCGCGGCAUAACAAACUGCCAUCCAUAAAUU --------......(((((((.........))...))))).........--------....(((((((..(((((((.....))))))).)))))))............... ( -21.10, z-score = -1.10, R) >droMoj3.scaffold_6500 22500210 100 + 32352404 -----------UCGGCGAGUCUCUCUGUGUCACUAGCGUCCUCUAUCUCGCGCCGUGAACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAUC-AUAAAUU -----------..((((............((((..(((..........)))...))))...(((((((..(((((((.....))))))).)))))))))))...-....... ( -27.00, z-score = -1.30, R) >droVir3.scaffold_12963 17454008 95 + 20206255 --------------GCGCCUCGCUGUGUCUCACUGGCGUCCUCUAUCUUUUGCCG--GACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAUC-AUAAAUU --------------((((......)))).....((((((((..((.....))..)--))).(((((((..(((((((.....))))))).))))))).))))..-....... ( -25.90, z-score = -1.63, R) >droGri2.scaffold_15252 14886953 98 + 17193109 ----------GUUGCCGACUAUCUGUGUCACUUUGGCGUCCUCUAUCUU-UGCUG--GACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAUC-AUAAAUU ----------......(((.......)))....((((((((..((....-))..)--))).(((((((..(((((((.....))))))).))))))).))))..-....... ( -28.20, z-score = -2.72, R) >droSec1.super_16 544797 105 + 1878335 CCCGACUUCUUGUAAAUAAAGUAUAUAUACUUUUUGCGUCCUCUAUCUUUC------AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU ...((......(((((..(((((....))))))))))..............------.((.(((((((..(((((((.....))))))).))))))).))..))-....... ( -21.10, z-score = -1.19, R) >droSim1.chr2L 12156865 105 + 22036055 ACCGACUUCUUGUAAAUAAAGUAUAUAUACUUUUUGCGUCCUCUAUCUUUC------AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAUC-AUAAAUU ...((......(((((..(((((....))))))))))..............------.((.(((((((..(((((((.....))))))).))))))).))..))-....... ( -21.10, z-score = -1.17, R) >consensus __CGACUUCUUGUAAAUACUGUAUAUAUACUUUUUGCGUCCUCUAUCUUUC______AGCUGUUUGUUUUUGUUGUGACAGGCGCGGCAUAACAAACUGCCAUC_AUAAAUU .............................................................(((((((..(((((((.....))))))).)))))))............... (-14.73 = -14.36 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:35:00 2011